Back to Journals » Journal of Inflammation Research » Volume 17
Circulating Exosomal microRNA Profiles Associated with Risk of Postoperative Recurrence in Chronic Rhinosinusitis with Nasal Polyps
Authors Chen S, Liu J, Feng Z, Zhou L, Cai Y, Jing Q
Received 12 June 2024
Accepted for publication 7 August 2024
Published 23 August 2024 Volume 2024:17 Pages 5619—5631
DOI https://doi.org/10.2147/JIR.S472963
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Tara Strutt
Siyuan Chen,1,2 Jia Liu,1,2 Zhili Feng,1,2 Liubo Zhou,1,2 Yuexiang Cai,1,2 Qiancheng Jing1,2
1Department of Otolaryngology Head and Neck Surgery, The Affiliated Changsha Central Hospital, Hengyang Medical School, University of South China, Changsha, Hunan, People’s Republic of China; 2Institute of Otolaryngology Head and Neck Surgery, Hengyang Medical School, University of South China, Changsha, Hunan, People’s Republic of China
Correspondence: Qiancheng Jing, Department of Otolaryngology Head and Neck Surgery, The Affiliated Changsha Central Hospital, Hengyang Medical School, University of South China, Changsha, Hunan, People’s Republic of China, Email [email protected]
Background: Exosomes carry various types of transcripts and serve as promising biomarkers for inflammatory diseases. However, the role of serum exosomal microRNAs (miRNAs) in chronic rhinosinusitis with nasal polyps (CRSwNP) is poorly clarified.
Methods: A prospective exploratory cohort of 10 CRSwNP patients was conducted, and the serum exosome samples were subjected to miRNA sequencing. Two independent prospective cohorts, consisting of 40 and 54 patients respectively, were recruited from different medical centers for validation. These cohorts were monitored for over two years, with postoperative recurrence serving as the primary outcome measure. The top 3 differentially exosomal miRNAs were validated in the serum samples, and their predictive values for recurrence were assessed.
Results: Eight CRSwNP patients completed the follow-up, comprising 4 non-recurrent cases and 4 recurrent cases. Distinctive profiles of serum exosomal miRNAs were identified between the two groups. In the first validation cohort, reverse transcription-polymerase chain reaction results indicated elevated serum exosomal miR-3174 and miR-6750-5p expressions, along with reduced miR-192-3p levels in the recurrence group compared to the non-recurrence group. Receiver operating characteristic (ROC) curves and Kaplan-Meier survival analysis demonstrated significant correlations between expressions of exosomal miR-3174 and miR-192-3p and the risk of postoperative recurrence. These findings were further validated in the second cohort, confirming the elevation of both miRNAs in the recurrence group and their associations with recurrence risk. Additionally, serum exosomal miR-3174 levels increased in recurrent cases compared to their baseline levels.
Conclusion: Circulating exosomal microRNA signatures may influence the risk of postoperative recurrence in CRSwNP patients. Serum exosomal elevated exosomal miR-3174 and decreased miR-192-3p were correlated with CRSwNP recurrence risk.
Keywords: exosome, chronic rhinosinusitis with nasal polyps, biomarker, recurrence
Introduction
Chronic rhinosinusitis with nasal polyps (CRSwNP) represents a prevalent and debilitating chronic inflammatory condition in the nasal cavity and sinuses.1,2 Despite advancements in medical therapies, and the widespread adoption of functional endoscopic sinus surgery (FESS) as the alternative surgical intervention for those patients who respond poorly to conservative treatments.3,4 Although the apparent potential of FESS can be advantageous for most patients, the effective management of this condition is impeded by the looming risk of postoperative recurrence.5,6 This challenge is exacerbated by the intricate etiology and pathogenic mechanisms underlying CRSwNP. The recurrence not only prompts the need for additional medical interventions and surgeries but also exerts a considerable toll on the patient’s overall quality of life.7,8 Postoperative recurrence frequently results in the return of disease symptoms, which can sometimes be more severe, causing increased distress for patients. Moreover, for individuals who do not respond adequately to medical treatments, revision surgery remains the sole option for symptom relief.9,10 Although several biomarkers, such as single-nucleotide polymorphisms,11 circulating metabolites,6 and circulating cytokines,12 have been associated with the recurrence of CRSwNP, none have been clinically validated or made available for routine use. Consequently, there is a critical need to delve into the identification of early predictive biomarkers for recurrent CRSwNP and to unravel the potential mechanisms underlying recurrence. Such efforts are pivotal for developing targeted treatment strategies and, ultimately, achieving the goal of individualized precision therapy.
In recent years, extracellular vesicles, particularly exosomes, have become crucial facilitators of communication between cells and potential repositories of disease-specific molecular information.13–15 Among the cargo transported by exosomes, microRNAs (miRNAs) stand out as promising candidates due to their regulatory roles in gene expression and their stability in various bodily fluids, including serum.16 Exosomes carrying miRNAs are involved in the underlying mechanisms of various diseases.16,17 Recently, accumulating evidence has indicated the considerable promise of circulating exosomal miRNAs as non-invasive biomarkers for disease prognosis and the prediction of recurrence.18,19 This potential has been demonstrated across various chronic inflammatory conditions, encompassing osteoarthritis,20 inflammatory bowel disease,21 chronic obstructive pulmonary disease,22 and allergic rhinitis.23 However, there is currently no reported information on the expression profile and role of circulating exosomal miRNAs in CRSwNP, as well as their potential as biomarkers for predicting postoperative recurrence.
This study aims to delineate the circulating exosomal miRNA profiles in CRSwNP patients through sequencing. It seeks to leverage the diagnostic and prognostic potential of serum exosomal miRNAs to identify distinctive molecular signatures linked to the risk of postoperative recurrence in CRSwNP patients, with validation conducted in two independent cohorts. The findings will contribute to the establishment of a clinically relevant, non-invasive tool that provides clinicians with valuable insights into the likelihood of postoperative recurrence, facilitating timely and targeted interventions to enhance long-term outcomes for individuals with CRSwNP.
Methods
Patients and Settings
This prospective study consists of one discovery cohort and two independent cohorts. The discovery cohort initially recruited 10 CRSwNP patients, and the first and second validation cohorts initially included 50 and 64 CRSwNP patients treated with FESS between March 2021 and May 2021. The including criteria are as following: (1) patients meeting the diagnostic criteria for CRSwNP as defined by the European Position Paper on Rhinosinusitis and Nasal Polyps 2012;24 (2) poor response to medication necessitating FESS; (3) age between 18 and 65 years; (4) no previous history of FESS.
Exclusion criteria: (1) incomplete clinical data, diagnoses of fungal sinusitis, allergic fungal rhinosinusitis, or sinonasal tumors; (2) recent use of oral corticosteroids, antibiotics within the last month; (3) concomitant acute inflammation or asthma; (4) age below 18 or above 65 years. The Ethics Committee of the Affiliated Changsha Central Hospital of Hengyang Medical School approved the research protocol (No. 2023097). All patients provided written informed consent.
Follow-Up and Definition of Recurrence
All CRSwNP patients underwent FESS performed by experienced surgeons, following consistent surgical standards. Postoperatively, patients received standard care, including nasal irrigation and steroid nasal spray. In cases of persistent symptoms, additional measures such as oral antibiotics and prednisone were administered as previously described.25,26 Postoperative follow-up appointments were performed to monitor disease progression. Recurrence was defined as the persistence of disabling symptoms, endoscopic indicators, or CT findings for at least 2 months despite the prescribed antibiotic and oral steroid rescue regimen.27,28 The minimum follow-up duration for all patients was 2 years, and they were divided into nonrecurrence and recurrence groups.
Clinical Data and Serum Sample Collection
Demographic and clinical data were collected from all subjects before their recruitment, including sex, age, body mass index (BMI), accompanying allergic rhinitis, asthma and aspirin intolerance, and Lund-MacKay and Lund-Kennedy scores. Blood samples from CRSwNP patients were collected using vacuum tubes containing a coagulant on the day of admission, usually 2–3 days before undergoing FESS. In the second validation cohort, blood specimens were collected from individuals who experienced recurrence, while those who did not experience recurrence provided blood samples again after a 2-year follow-up. All harvested samples were immediately centrifuged at 3000 g for 10 min to isolate serum samples. The collected serum specimens were kept at −80 °C until analysis.
Serum Exosome Isolation and Identification
The serum exosomes were isolated with the Total Exosome Isolation Reagent kit (Invitrogen®, USA), and then subjected to either identification or RNA extraction. After purifying the exosomes, transmission electron microscopy (TEM) was utilized to observe their structure and morphology following previously described protocols.22 The particle size and size distribution of exosomes were analyzed by dynamic light scattering. Western bolting was performed to detect the protein expression of the specific exosome markers in both exosome samples and serum without exosome samples. The specific markers primary antibodies, including CD81 (Affinity, China) and CD63 (Affinity, China), and secondary antibodies (Affinity, China) were used.
RNA Extraction and RNA-Sequencing
RNA was extracted from exosomes obtained from CRSwNP patients, and the concentration and purity were assessed. Following RNA extraction, a sequencing library was prepared using the NEB Multiplex Small RNA Library Prep Set for Illumina (Illumina, USA) according to established protocols outlined in previous studies.23,29 This process involved reverse transcription, cDNA synthesis, adapter addition, and sequencing primer ligation. Following these steps, the sequencing library was subjected to 50-cycle sequencing on the Illumina NextSeq 500 platform (Illumina, USA).
Data Analysis and Target Prediction
In the discovery cohort, 8 CRSwNP patients completed the follow-up, including 4 cases without recurrence and 4 cases with recurrence. The raw sequencing data received stringent quality control and subsequent analysis using the statistical R package. Differentially expressed miRNAs were identified based on a fold change (FC) ≥1.5 or FC ≤0.67 and a P-value <0.05, comparing the non-recurrence and recurrence groups. Heatmap analysis and volcano plots were generated to visually represent the differentially expressed miRNAs. Pathway enrichment analysis was conducted to discern the enriched target genes of the differentially expressed miRNAs.
Validation of miRNA
To corroborate the sequencing findings, we selected the three most dysregulated exosomal miRNAs for validation using reverse transcription-quantitative polymerase chain reaction (RT-PCR). During this procedure, RNA extraction from exosomes was utilized as a template for reverse transcription. The cDNA synthesis was carried out using total RNA and the Mir-X miRNA First-Strand Synthesis Kit (Takara, China) following the provided instructions. Following this, quantitative PCR was employed to amplify and quantify the cDNA molecules associated with the target miRNAs, using the miRNAs primer outlined in Table S1. The relative expression levels of miRNAs were normalized to the external control cel-miR-39 as previously described.23,29
Statistical Analysis
Variables with a normal distribution are reported as mean ± SD and analyzed using Student’s t-test. For variables lacking a normal distribution, median and interquartile ranges (IQRs) are provided, and the Mann–Whitney U-test is utilized for comparisons. Categorical data are shown as frequencies and percentages, and compared with the chi-square test. Receiver operating characteristic (ROC) curves are employed to evaluate the predictive values of serum exosomal miRNAs for recurrence, the area under the curve (AUC) and 95% confidence interval (CI) were recorded to evaluate the predictive ability. CRSwNP patients are stratified into high and low-miRNA groups based on the median values of serum exosomal miRNA levels. Kaplan-Meier survival analysis is conducted to analyze their associations with the risk of postoperative recurrence. Data analysis is performed using SPSS 21.0 (SPSS, USA) and GraphPad Prism 7.0 (GraphPad, USA). A significance level of 0.05 is applied for all tests, with p-values below this threshold considered statistically significant.
Results
Identification of Exosomes
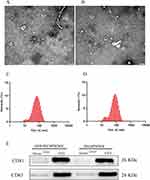
Following a two-year follow-up, 4 CRSwNP patients were assigned to the non-recurrence group, while 4 patients were categorized into the recurrence group. TEM images displayed the typical round cup-shaped morphology of exosomes collected from both groups (Figure 1A and B). Nanoparticle tracking analysis results illustrated particle diameters ranging from 80 to 100 nm (Figure 1C and D). Additionally, WB images demonstrated significant expressions of CD81 and CD63 in the detected exosome samples, with sparse expression observed in serum samples devoid of exosomes (Figure 1E). These findings collectively confirm the successful collection of exosomes from the two groups.
Serum Exosomal miRNA Profiles Between the Non-Recurrence and Recurrence Groups
The sequencing results, as illustrated by heat maps and volcano plots in Figure 2A and B, unveiled distinct serum exosomal miRNA profiles distinguishing the non-recurrence and recurrence groups. A total of 818 serum exosomal miRNA were detected between the two groups. Among them, 74 miRNAs exhibited differential expression in the recurrence group compared to the non-recurrence group, with 46 being up-regulated and 28 down-regulated. To elucidate the biological significance of exosomal miRNAs in CRSwNP, we predicted potential target genes for the differentially expressed miRNAs. The KEGG pathway analysis indicated that these miRNAs were prominently involved in pathways related to bacterial invasion of epithelial cells and the TGF-beta signaling pathway (Figure 2C and D).
Validation of Exosomal miRNA Profiles in Validation Cohorts
The top 3 dysregulated serum exosomal miRNAs were selected for validation in the first validation cohort. The detailed parameters of these miRNAs are listed in Table 1. This first validation cohort comprised 40 CRSwNP patients who completed a two-year follow-up, with 25 patients categorized as non-recurrent and 15 as recurrent. Table 2 details the demographic and clinical data between the two groups. The RT-PCR results in Figure 3 showed that serum exosomal miR-4657, miR-6750-5p, and miR-6750-5p levels were increased, while miR-192-3p levels were reduced in the recurrence group compared to the non-recurrence group. ROC curves demonstrated that serum exosomal miR-6750-5p, miR-3174, and miR-192-3p levels exhibited predictive values for CRSwNP recurrence (Figure 4A–F). Additionally, Kaplan-Meier survival analysis suggested that serum exosomal miR-3174 and miR-192-3p levels were associated with the risk of postoperative recurrence (Figure 4G–L).
 |
Table 1 The Parameters of Top 3 Up-Regulated and Down-Regulated miRNAs |
 |
Table 2 Demographic Characteristics of CRSwNP Patients in the First Validation Cohort |
To further confirm the associations between potential serum exosomal miRNAs and postoperative recurrence, we assessed the levels of serum exosomal miR-3174 and miR-192-3p in the second validation cohort from another medical center. This cohort included 54 CRSwNP patients, with 35 non-recurrent patients and 19 recurrent patients after a two-year follow-up. Table 3 provides the demographic and clinical data between the two groups. The RT-PCR results in Figure 5A and B revealed that miR-3174 was increased, and miR-192-3p was decreased in the serum exosomes in the recurrence group compared to the non-recurrence group. ROC and Kaplan-Meier survival analysis suggested that serum exosomal miR-3174 and miR-192-3p levels were associated with the risk of postoperative recurrence (Figure 5C–F). Moreover, our findings indicated that serum exosomal miR-3174 and miR-192-3p levels were significantly enhanced when CRSwNP patients experienced postoperative recurrence compared to the baseline levels, with no significant change observed in non-recurrent cases between baseline and 2 years post-FESS levels (Figure 6). These results imply that elevated serum exosomal miR-3174 may contribute to the recurrent mechanisms of CRSwNP.
 |
Table 3 Demographic Characteristics of CRSwNP Patients in the Second Validation Cohort |
Discussion
The pathological mechanisms of CRSwNP are poorly clarified, and the high rate of recurrence poses a significant clinical challenge due to its high tissue heterogeneity. Exploring early predictors of recurrent CRSwNP and clarifying the underlying recurrent mechanisms are current hot research topics and challenges. Although previous studies have identified various objectively abnormal expressions of biomarkers closely associated with the risk of CRSwNP recurrence, such as serum metabolites,6 nasal microbiota,30 and tissue eosinophilia,28 the accuracy of these biomarkers needs further improvement before being applied in clinical practice.
Exosomes, as standout molecules in current research, can express miRNAs in the serum.22,31 Previous studies have confirmed that various diseases exhibit specific serum exosomal miRNA expression profiles and multiple miRNAs have been identified with potential capabilities for disease diagnosis and prognosis assessment.32,33 Although several previous studies have emphasized the involvement of tissue circRNA/miRNA ceRNA networks in the pathological mechanisms of CRSwNP,34 there is currently a lack of research on the expression and mechanisms of exosome-carried miRNAs in CRSwNP. Despite a recent study discovering differences in the exosomal miRNA expression profile in nasal lavage fluid between chronic rhinosinusitis patients and healthy controls, the clinical significance and specific mechanisms of the associated miRNAs were not elucidated.35 Therefore, exploring the associations between serum exosomal miRNA expression profiles and postoperative recurrence in CRSwNP, and clarifying the intricate interplay between these small RNA molecules and the molecular pathways implicated in CRSwNP pathogenesis provides a unique opportunity to uncover novel insights into the mechanisms governing postoperative recurrence.
In this prospective study, we first analyzed serum exosomal miRNA profiles in CRSwNP patients and found that recurrent CRSwNP patients exhibited distinctive baseline miRNA profiles in comparison with non-recurrent cases. The candidate serum exosomal miRNAs were subsequently validated in two independent cohorts from different medical centers, confirming the predictive values of serum exosomal miR-3174 and miR-192-3p for postoperative recurrence in CRSwNP. Intriguingly, miR-3174 was significantly increased in the serum exosomes of CRSwNP patients who suffered a recurrence in comparison with their baseline levels. These results suggest that serum exosomal miRNA profiles affected the risk of recurrence of CRSwNP, and miR-3174 might be involved in its recurrent mechanisms.
We demonstrated that serum exosomal miR-3174 levels correlated with the risk of postoperative recurrence in CRSwNP patients, and its level change was involved in the process of recurrence. KEGG pathway analysis showed that several target genes of miR-3174 were involved in the bacterial invasion of epithelial cells and the TGF-beta signaling pathways. It is well established that epithelial cell bacterial invasion and TGF-beta signaling activation were crucial in the immunopathological mechanisms of CRSwNP and associated with the recurrence.36–38 Bacterial invasion, injury, and inflammation in nasal mucosal epithelial cells can significantly impact the barrier function of nasal mucosal epithelium, hindering its repair.39,40 Additionally, the accumulation of TGF- beta 1 promotes tissue remodeling and epithelial-mesenchymal transition (EMT) in nasal mucosal epithelium. These pathological changes have been confirmed to be closely associated with the postoperative recurrence of CRSwNP.41,42 Recent studies have identified miR-3174 as an oncogene capable of regulating autophagy and apoptosis in tumor cells, promoting interstitial invasion of tumor cells.43,44 Another study has confirmed that miR-3174 can facilitate the differentiation of tumor stromal cells, mediating the occurrence of EMT in liver cancer tissues.45 Combining the current research findings with existing literature, we speculate that miR-3174 carried by extracellular vesicles in serum may influence the differentiation of stromal cells in the nasal mucosa, promoting EMT and tissue remodeling, thereby participating in the postoperative recurrence process of CRSwNP. However, the relevant regulatory mechanisms require further elucidation.
Another interesting finding was that serum exosomal miR-192-3p levels were reduced in the recurrent CRSwNP patients, and its levels were negatively correlated with the risk of postoperative recurrence. Currently, existing research primarily focuses on the role of miR-192-3p in tumors, revealing that its aberrant expression levels are closely associated with tumorigenesis and biological behavior.46,47 A recent publication demonstrated that miR-192-3p was dysregulated in the synchronous colorectal adenoma tissues and associated with tissue immune cell profile, including dendritic cells, eosinophils, and macrophages.48 Hu et al49 found that miR-192-3p could target IGF2 and regulate macrophage polarization, which contributed to the etiopathogenesis of hepatocellular carcinoma. Accordingly, the immune microenvironment changes mediated by immune cells within the mucosal tissue, including macrophage M2 polarization and eosinophil recruitment are crucial mechanisms in mediating the postoperative recurrence of CRSwNP.50,51 Combining the results of this study with existing literature, we speculate that serum exosomal miR-192-3p may influence the functionality of immune cells in nasal polyp tissue, including the regulation of macrophage M2 polarization and eosinophil recruitment, thereby affecting tissue inflammation development and postoperative recurrence. However, specific mechanisms await further confirmation.
Our study has several limitations. Firstly, it is a single-center prospective study with a limited sample size and absence of healthy controls, and the predictive values of serum exosomal miRNAs should be validated in a larger, multi-center cohort study. Secondly, we focused solely on the analysis of baseline serum exosomal miRNAs in CRSwNP patients and their predictive values for postoperative recurrence, without comparing them between CRSwNP patients and healthy controls. Thirdly, we did not undertake mechanistic studies to elucidate the role of candidate miRNAs in the recurrence mechanism of CRSwNP. It is crucial to further investigate the underlying mechanisms to understand how these miRNAs contribute to the recurrence process and to explore their potential roles in disease progression and therapeutic targeting.
Conclusions
In this prospective study, we confirmed that serum exosomal miRNA profiles were associated with the risk of postoperative recurrence in CRSwNP patients. The discovery-validation results highlighted that serum exosomal elevated exosomal miR-3174 and decreased miR-192-3p were correlated with CRSwNP recurrence risk, suggesting they were potential biomarkers for predicting postoperative recurrence.
Ethics and Consent Statements
This study was approved by the Ethics Committee of the Affiliated Changsha Central Hospital of Hengyang Medical School approved the research protocol (No. 2023097). All patients provided written informed consent. This study complies with the Declaration of Helsinki.
Funding
This research was supported by the Changsha Municipal Natural Science Foundation Project (No. kq2208444), the Changsha Municipal Health Commission Fund Project (No. SB2023-226).
Disclosure
The authors declare no conflicts of interest in preparing this article.
References
1. Boechat JL, Sousa-Pinto B, Delgado L, et al. Biologicals in severe chronic rhinosinusitis with nasal polyps: translation to clinical practice while waiting for head-to-head studies. Rhinology. 2023;61(3):283–286. doi:10.4193/Rhin22.436
2. Boscke R, Heidemann M, Bruchhage KL. Dupilumab for chronic rhinosinusitis with nasal polyps: real-life retrospective 12-month effectiveness data. Rhinology. 2023;61(3):203–213. doi:10.4193/Rhin22.469
3. Lee SE, Amin N, Mannent LP, et al. The relationship of sinus opacification, olfaction and dupilumab efficacy in patients with CRSwNP. Rhinology. 2023;61(6):531–540. doi:10.4193/Rhin22.220
4. Sedaghat AR, Fokkens WJ, Lund VJ, et al. Consensus criteria for chronic rhinosinusitis disease control: an international delphi study. Rhinology. 2023;61(6):519–530. doi:10.4193/Rhin23.335
5. Bai J, Huang JH, Price CPE, et al. Prognostic factors for polyp recurrence in chronic rhinosinusitis with nasal polyps. J Allergy Clin Immunol. 2022;150(2):352–361.e357. doi:10.1016/j.jaci.2022.02.029
6. Xie S, Zhang C, Xie Z, et al. Serum metabolomics identifies uric acid as a possible novel biomarker for predicting recurrence of chronic rhinosinusitis with nasal polyps. Rhinology. 2023;61(6):541–551. doi:10.4193/Rhin23.236
7. Kemp P, van der Lans RJL, Otten JJ, et al. Hypereosinophilia during dupilumab treatment in patients with chronic rhinosinusitis with nasal polyps. Rhinology. 2024;62(2):202–207. doi:10.4193/Rhin23.357
8. Shen KH, Jiang JY, Hsu PY, et al. Can serum IgE or blood eosinophil count predict postoperative oral corticosteroid response in chronic rhinosinusitis with nasal polyps? Rhinology. 2024;62:192–201.
9. Xie Y, Liang F, Zhou L, et al. Correlation among post-surgery recurrence of CRSwNP and TCM syndromes and tissue inflammatory cell infiltration type: a study protocol. Syst Rev. 2024;13(1):145. doi:10.1186/s13643-024-02562-9
10. Zhou Y, Feng Z, Wen J, et al. Aberrant expressions of TAM receptors are associated with postoperative recurrence in chronic rhinosinusitis with nasal polyps. European Archives of Oto-Rhino-Laryngology: Official journal of the European Federation of Oto-Rhino-Laryngological Societies (EUFOS): Affiliated with the German Society for Oto-Rhino-Laryngology Head and Neck Surgery. 2024;281(6):3005–3015. doi:10.1007/s00405-024-08450-1
11. Antonino M, Nicolò M, Jerome Renee L, et al. Single-nucleotide polymorphism in chronic rhinosinusitis: a systematic review. Clin Otolaryngol. 2022;47:14–23.
12. Wang G, Zheng H, Chen X, et al. Exploration of predictive biomarkers for postoperative recurrence in chronic rhinosinusitis with nasal polyps based on serum multiple-cytokine profiling. Mediators Inflamm. 2022;2022:1061658. doi:10.1155/2022/1061658
13. Tenchov R, Sasso JM, Wang X, et al. Exosomes─nature’s lipid nanoparticles, a rising star in drug delivery and diagnostics. ACS nano. 2022;16:17802–17846.
14. An J, Park H, Kim J, et al. Extended-Gate Field-Effect Transistor Consisted of a CD9 aptamer and mxene for exosome detection in human serum. ACS Sens. 2023;8(8):3174–3186. doi:10.1021/acssensors.3c00879
15. Ye H, Wang K, Zhao J, et al. In situ sprayed nanovaccine suppressing exosomal PD-L1 by golgi apparatus disorganization for postsurgical melanoma immunotherapy. ACS nano. 2023;17(11):10637–10650. doi:10.1021/acsnano.3c01733
16. Fan W, Pang H, Shi X, et al. Plasma-derived exosomal mRNA profiles associated with type 1 diabetes mellitus. Front Immunol. 2022;13:995610. doi:10.3389/fimmu.2022.995610
17. Kim DH, Park H, Choi YJ, et al. Identification of exosomal microRNA panel as diagnostic and prognostic biomarker for small cell lung cancer. Biomarker Res. 2023;11(1):80. doi:10.1186/s40364-023-00517-1
18. Singh T, Kaushik M, Mishra LC, et al. Exosomal miRNAs as novel avenues for breast cancer treatment. Front Genetics. 2023;14:1134779. doi:10.3389/fgene.2023.1134779
19. Chang M, Jiang S, Guo X, et al. Exosomal RNAs in the development and treatment of pituitary adenomas. Front Endocrinol. 2023;14:1142494. doi:10.3389/fendo.2023.1142494
20. Fan WJ, Liu D, Pan LY, et al. Exosomes in osteoarthritis: updated insights on pathogenesis, diagnosis, and treatment. Front Cell Develop Biol. 2022;10:949690. doi:10.3389/fcell.2022.949690
21. Bauer KM, Nelson MC, Tang WW, et al. CD11c+ myeloid cell exosomes reduce intestinal inflammation during colitis. JCI Insight. 2022;7. doi:10.1172/jci.insight.159469
22. Wang F, Yang B, Qiao J, et al. Serum exosomal microRNA-1258 may as a novel biomarker for the diagnosis of acute exacerbations of chronic obstructive pulmonary disease. Sci Rep. 2023;13(1):18332. doi:10.1038/s41598-023-45592-4
23. Cao S, Wu H, Niu Y, et al. Circulating exosomal has-miR-24-3p and has-miR-128-3p reflect early efficacy of sublingual immunotherapy in allergic rhinitis. Int Immunopharmacol. 2023;124(Pt A):110822. doi:10.1016/j.intimp.2023.110822
24. Fokkens WJ, Lund VJ, Mullol J, et al. European position paper on rhinosinusitis and nasal polyps 2012. Rhinol Suppl. 2012;23(3):1–298.
25. Li JX, Wang ZZ, Zhai GT, et al. Untargeted metabolomic profiling identifies disease-specific and outcome-related signatures in chronic rhinosinusitis. J Allergy Clin Immunol. 2022;150(3):727–735.e726. doi:10.1016/j.jaci.2022.04.006
26. Renteria AE, Maniakas A, Pelletier A, et al. Utilization of transcriptomic profiling to identify molecular markers predicting successful recovery following endoscopic sinus surgery for chronic rhinosinusitis. Otolaryngology--Head and Neck Surgery: Official Journal of American Academy of Otolaryngology-Head and Neck Surgery. 2023;169(6):1662–1673. doi:10.1002/ohn.482
27. Wang T, Chen Y, Gao R, et al. Overexpression of AXL on macrophages associates with disease severity and recurrence in chronic rhinosinusitis with nasal polyps. Int Immunopharmacol. 2023;121:110449. doi:10.1016/j.intimp.2023.110449
28. Wu PW, Chiu CH, Huang YL, et al. Tissue eosinophilia and computed tomography features in paediatric chronic rhinosinusitis with nasal polyps requiring revision surgery. Rhinology. 2023;61:246–254.
29. Jiang S, Xie S, Fan R, et al. Exosomes derived hsa-miR-4669 as a novel biomarker for early predicting the response of subcutaneous immunotherapy in pediatric allergic rhinitis. J Inflamm Res. 2022;15:5063–5074. doi:10.2147/JIR.S379414
30. Zhao Y, Chen J, Hao Y, et al. Predicting the recurrence of chronic rhinosinusitis with nasal polyps using nasal microbiota. Allergy. 2022;77(2):540–549. doi:10.1111/all.15168
31. Utkarsh K, Kumar A, Khan A, et al. Circulating and non-circulating proteins and nucleic acids as biomarkers and therapeutic molecules in ovarian cancer. Genes Dis. 2023;10:1005–1018.
32. Yang Y, Huang H, Li Y. Roles of exosomes and exosome-derived miRNAs in pulmonary fibrosis. Front Pharmacol. 2022;13:928933. doi:10.3389/fphar.2022.928933
33. Qian W, Huang L, Xu Y, et al. Hypoxic ASCs-derived exosomes attenuate colitis by regulating macrophage polarization via miR-216a-5p/HMGB1 axis. Inflamm Bowel Dis. 2023;29(4):602–619. doi:10.1093/ibd/izac225
34. Sun Q, Liu Z, Xu X, et al. Identification of a circRNA/miRNA/mRNA ceRNA network as a cell cycle-related regulator for chronic sinusitis with nasal polyps. J Inflamm Res. 2022;15:2601–2615. doi:10.2147/JIR.S358387
35. Cha S, Seo EH, Lee SH, et al. MicroRNA expression in extracellular vesicles from nasal lavage fluid in chronic rhinosinusitis. Biomedicines. 2021;9:471.
36. Szaleniec J, Gibała A, Hartwich P, et al. Challenging the gold standard: methods of sampling for microbial culture in patients with chronic rhinosinusitis. European archives of oto-rhino-laryngology: official journal of the European Federation of Oto-Rhino-Laryngological Societies (EUFOS): affiliated with the German Society for Oto-Rhino-Laryngology. Head and Neck Surg. 2021;278:4795–4803.
37. Li XL, Feng QM, Yang HN, et al. p120 regulates E-cadherin expression in nasal epithelial cells in chronic rhinosinusitis. Rhinology. 2022;60(4):270–281. doi:10.4193/Rhin21.276
38. Xu X, Seet JE, Yap QV, et al. Latent class analysis of structured histopathology in prognosticating surgical outcomes of chronic rhinosinusitis with nasal polyps in Singapore. Rhinology. 2023;61:358–367.
39. Liao S, Huang Y, Zhang J, et al. Vitamin D promotes epithelial tissue repair and host defense responses against influenza H1N1 virus and Staphylococcus aureus infections. Respir Res. 2023;24(1):175. doi:10.1186/s12931-023-02477-4
40. Xiao Q, Wang H, Song J, et al. Impaired local Vitamin D3 metabolism contributes to IL-36g overproduction in epithelial cells in chronic rhinosinusitis with nasal polyps. Rhinology. 2024;62(2):236–249. doi:10.4193/RhinRhin23.123
41. Porras-Gonzalez C, Palacios-Garcia JM, Sanchez-Gomez S, et al. Transcriptional analysis of nasal polyps fibroblasts reveals a new source of pro-inflammatory signaling in CRSwNP. Rhinology. 2023;61:180–189.
42. Jo S, Lee SH, Jo HR, et al. Eosinophil-derived TGFβ1 controls the new bone formation in chronic rhinosinusitis with nasal polyps. Rhinology. 2023;61(4):338–347. doi:10.4193/Rhin22.439
43. Zhang DL, Yang N. MiR-3174 functions as an oncogene in rectal cancer by targeting PCBD2. Eur. Rev. Med. Pharmacol. Sci. 2019;23(6):2417–2426. doi:10.26355/eurrev_201903_17388
44. Li QY, Shen JQ, Li JH, et al. LINC00958/miR-3174/PHF6 axis is responsible for triggering proliferation, migration and invasion of endometrial cancer. Eur. Rev. Med. Pharmacol. Sci. 2021;25:6853–6861.
45. Wang Q, Yang X, Zhou X, et al. MiR-3174 promotes proliferation and inhibits apoptosis by targeting FOXO1 in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2020;526(4):889–897. doi:10.1016/j.bbrc.2020.03.152
46. Chen Z, Lin W, Cai Q, et al. A large-scale microRNA transcriptome-wide association study identifies two susceptibility microRNAs, miR-1307-5p and miR-192-3p, for colorectal cancer risk. Human Molecular Genetics. 2023;33(4):333–41.
47. Li F, Deng Y, Zhang S, et al. Human hepatocyte-enriched miRNA-192-3p promotes HBV replication through inhibiting Akt/mTOR signalling by targeting ZNF143 in hepatic cell lines. Emerging Microbes Infect. 2022;11:616–628.
48. Ng L, Li X, Wan TM, et al. Investigation of miRNA dysregulation and association with immune cell profile during malignant transformation of colorectal cells. Eur J Surg Oncol. 2022;48(1):245–252. doi:10.1016/j.ejso.2021.09.016
49. Hu C, Li X, Sui Y, et al. Dicer deletion in hepatocytes promotes macrophages M1 polarization through dysregulated miR-192-3p/IGF2 in non-alcoholic steatohepatitis and hepatocellular carcinoma. Cancer Gene Ther. 2022;29(8–9):1252–1262. doi:10.1038/s41417-022-00432-x
50. Rizzi A, Gammeri L, Cordiano R, et al. Therapeutic strategies to prevent the recurrence of nasal polyps after surgical treatment: an update and in vitro study on growth inhibition of fibroblasts. J Clin Med. 2023;12:2841.
51. Sima Y, Zhao Y, Wang X, et al. Precision medicine in chronic rhinosinusitis - using endotype and endotype-driven therapeutic options. Expert Rev Clin Immunol. 2023;19:1–10.
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.