Back to Journals » Journal of Inflammation Research » Volume 17
Diagnosis and Molecular Characterization of Potential RNA Binding Protein Involved in the Pathogenesis of Liver Ischemia Reperfusion Injury
Received 19 May 2024
Accepted for publication 16 July 2024
Published 22 July 2024 Volume 2024:17 Pages 4881—4893
DOI https://doi.org/10.2147/JIR.S468828
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Weiju Lai,1,* Jiajian Yu,2,* Diguang Wen3
1Central Laboratory, Chongqing FuLing Hospital, School of Medicine, Chongqing University, Chongqing, People’s Republic of China; 2Department of Hepatobiliary, Chongqing Fuling Hospital, School of Medicine, Chongqing University, Chongqing, People’s Republic of China; 3Second Affiliated Hospital of Chongqing Medical University, Chongqing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Diguang Wen, Email [email protected]; [email protected]
Background: Liver ischemia-reperfusion is one of the common complications after liver surgery. Uncontrolled liver ischemia-reperfusion will lead to many serious consequences such as surgical failure. It is an urgent clinical problem to search for diagnostic markers and explore its potential pathogenesis.
Methods: In this study, we focus on 1411 candidate RNA binding protein. Through several GEO (Gene Expression Omnibus) online datasets, we construct a diagnostic model and perform interactive validation. We evaluate the efficacy of the prognostic model. Using bioinformatics methods, we predicted the relevant signaling pathways of liver ischemia-reperfusion and key genes. We also evaluated the association of RNA binding protein with immune cell infiltration. Single cell sequencing datasets were used to explore the expression profiles of key genes at the single cell level. Machine learning algorithm is used to predict key gene RNA binding domains.
Results: ROC (Receiver Operating Characteristic) and DCA (Decision Curve Analysis) curves showed that the above diagnostic model had good and stable diagnostic efficacy and clinical practicability. We identified three key genes (BTG2, CCNL1 and DNAJB1) in liver ischemia-reperfusion. DNAJB1, BTG2 and CCNL1 are mainly expressed in immune cells such as macrophages and T cells, and are closely related to inflammatory pathways such as TNF-α, highlighting their importance in hepatic ischemia reperfusion. We identified RNA-binding domains of the above three genes. We found that the expression of DNAJB1, CCNL1 and BTG2 in the ischemia-reperfusion group were significantly higher than those in the sham operation group.
Conclusion: Our study revealed the importance of the candidate RNA binding protein in liver ischemia reperfusion injury and provided new insights into the therapeutic of hepatic ischemia-reperfusion injury.
Keywords: RBPs, liver ischemia reperfusion injury, bioinformatics
Introduction
Liver ischemia-reperfusion injury (I/RI) refers to the phenomenon that the liver tissue recovers blood perfusion after ischemia for a period of time, which not only cannot restore its function and structure, but aggravate its dysfunction and structural damage.1,2 Hepatic ischemia-reperfusion injury is commonly seen in hemorrhagic shock, hepatectomy and liver transplantation.3,4 Existing studies have shown that severe uncontrolled hepatic I/RI can lead to many dangerous clinical outcomes, including surgical failure, graft dysfunction, immune rejection, and even patient death.5,6 Liver I/RI is mainly characterized by inflammatory storm and death of a large number of liver cells, which ultimately impair liver function.7 RNA-binding proteins (RBPs) are a class of important proteins that have the function of RNA binding and regulate RNA metabolism in cells.8,9 RBPs have been found to play a key role in regulating liver ischemia-reperfusion, inflammatory response, and cell death, and are highly plastic, making them promising targets for clinical intervention.10,11 However, previous studies only focused on the role of classical RNA-binding proteins in hepatic ischemia-reperfusion injury.8,9,12 Jin et al have developed a new machine learning algorithm for identifying RNA-binding proteins with statistical accuracy of more than 90%, providing a new means for writing RNA binding maps.13 However, the role and function of these novel candidate RBPs in liver ischemia-reperfusion injury remain unclear, so this study conducted a systematic biogenic analysis of these RBPs to provide a new perspective for liver ischemia-reperfusion injury.
Methods
Differential Analysis of Expression of Candidate RNA-Binding Proteins
The list of RNA-binding proteins was identified by reference to previous machine learning algorithms. The liver ischemia-reperfusion injury dataset is downloaded from the GEO database (GSE12720 42 samples; GSE14951 15 samples; GSE151648 80 samples). The cutoff value for gene difference analysis was an absolute logFc value greater than 0.26 and a P value less than 0.05.
Construct Diagnostic Model
The intersection of differential genes in GSE12720 and GSE14951 datasets was selected as candidate genes for downstream analysis. The GSE12720 dataset was used to construct the diagnostic model. First, we construct LASSO regression models in order to screen for important genes. Next, two-way logistics regression analysis was used to construct the diagnostic model. The GSE14951 and GSE151648 datasets was used to validate the diagnostic model. The GEO-meta dataset contains three datasets (GSE12720, GSE14951 and GSE151648).
Kyoto Encyclopedia of Genes and Genomes (KEGG), Metascape and Gene Ontology (GO) Analysis
KEGG, Metascape and GO analysis were used to identify liver I/RI related signaling pathways. The GEO-meta dataset was used for gene difference analysis, and the cutoff value was logFc absolute value greater than 0.585 and P value less than 0.05. KEGG and GO were implemented to identify I/RI related signaling pathways using R software. Metascape analysis is the uploading of differential gene sets to an online website (https://metascape.org/gp/index.html#/reportfinal/ttf_17jw5) for analysis.14
Gene Correlation Analysis
R software is used for gene correlation analysis, Pearson’s test is used for gene correlation analysis, and correlation greater than 0.45 is used as cutoff values. Cytoscape_v3.9.0 is used to build correlation graphs.
Single Cell Sequencing Analysis
Single cell dataset was used GSE171539 dataset. In short, the UMAP function performs single-cell dimensionality reduction. Single cell Cluster annotation was performed based on each cluster marker, and the expression of BTG2, CCNL1 and DNAJB1 in each cell population was analyzed.
Liver Ischemia-Reperfusion Model in Mice
Male C57BL/6 mice aged 10 ~ 12 weeks were purchased from the Laboratory Animal Center of Chongqing Medical University. This model construction method is based on our previous research.15 The animals were anesthetized and the blood supply to the left/middle liver lobe was interrupted using a noninvasive clamp, resulting in 70% liver ischemia. After 90 minutes of ischemia, the forceps were removed. The sham control group had the same procedure, but no blood vessels were blocked.
Immunohistochemistry
For immunohistochemical analysis, liver sections were rehydrated, underwent an antigen removal procedure, and then incubated overnight at 4°C with primary antibodies of BTG2 (proteintech, China), CCNL1 (Affinity, USA), and DNAJB1 (Affinity, USA). On the second day, incubation containing HRP secondary antibody was conducted at room temperature for 1 hour, and then DAB staining was performed. Hematoxylin is used to stain the nucleus. Then ethanol gradient dehydration, xylene transparent. The H-score (Histochemistry-score) is used to evaluate the intensity of immunohistochemistry.
Characteristic of Immune Infiltration
The CIBERSORT algorithm was applied to comprehensively assess the immunological characteristics of all samples included in the study. The correlation between the number of immune cells and RBPs expression was conducted by Pearson’s test.
RNA Binding Protein Domain Analysis
Deep-learning model HydRA is used for RNA binding protein domains analysis.13 UniProt is used to download protein structure of key genes files that predict being AlphaFold. Pymol 1.8.6 is used to map protein structures.
Statistical Analysis
SPSS 24.0 and GraphPad Prism 8.0 (GraphPad, La Jolla, CA) were used for statistical analyses. The measured data are represented as means ± SEM. Two-tailed Student’s t-test was conducted to compare quantitative data. The correlation test was conducted by Pearson’s test. P < 0.05 was considered statistically significant.
The analysis flow chart is shown in Figure 1A.
Results
Differential Expression Analysis of Candidate RNA Binding Proteins
We obtained 1411 RNA-binding proteins based on previous studies (Table S1). The difference analysis of a single dataset is prone to bias, so we selected two GEO online datasets for differential expression analysis respectively, and the intersection was used to obtain candidate genes. We carried out gene difference analysis using GSE12720 and GSE14951 datasets. In GSE12720, we identified 12 up-regulated genes and 1 down-regulated gene in I/RI (Figure 1B). In GSE14951, we identified 43 up-regulated genes and 3 down-regulated genes in I/RI (Figure 1B). After intersection, we finally obtained 9 RBPs up-regulated in I/RI for subsequent analysis which a (Figure 1C).
Establish and Validate Diagnostic Models
Establishing a stable and reliable diagnostic model can help us to intervene in the early stage of liver ischemia-reperfusion patients. Through lasso regression analysis, we obtained six important RBPs (DNAJB1, PP1R15A, BTG2, HSPA6, CCNL1, NAMPT) (Figure 2A). Further logistics regression analysis, we identified 3 important features and built a diagnostic model (Risk Scores=204.37*DNAJB1+BTG2*1398.65+1379.64*CCNL1). The ROC curve shows an AUC value of 1.000, which indicates that the model has good diagnostic efficacy in GSE12720 dataset (Figure 2B). In the external data set, the results show that AUC values are all higher than 0.9 (AUC value 1.000 in GSE14951; AUC value 0.946 in GSE151648; AUC value 0.915 in GSE-meta), indicating that the model has good stability (Figure 2C). DCA curve shows that this model has good safety interval and good clinical value (Figure 2D).
Molecular Signal Function Analysis
To identify potential liver I/RI signaling pathways, we performed differential expression analysis in a GEO-meta dataset and obtained 582 differentially expressed genes (Figure 3A and Table S2). GO analysis showed that the differential pathways were enriched in response to molecule of bacterial origin, signaling receptor activator activity and other pathways (Figure 3B and Table S3). METASCAPE analysis showed that differential genes were enriched in inflammation, infection and other pathways (Figure 3C). KEGG analysis showed that differential genes were enriched in IL-17, TNF-α and JAK-STAT pathways (Figure 3D and Table S4). These results support the critical role of inflammatory pathways in liver I/RI.16 Interestingly, GSEA analysis showed that the functions of BTG2, CCNL1, and DNAJB1 were all enriched in inflammatory pathways such as IL-17 and MAPK, supporting the potential core functions of these three molecules in hepatic I/RI (Figure 3E).
Correlation Analysis of Immune Cell Infiltration
A large number of inflammatory immune cell aggregation and functional transformation are the core features of inflammatory diseases such as I/RI.17 CIBERSORT analysis showed significant enrichment of monocytes and neutrophils after liver I/RI, and decreased infiltration of M2-type macrophages, which was consistent with previous cognition (Figure 4A and B).18 Further correlation analysis showed that BTG2, CCNL1 and DNAJB1 were closely related to the activation of mast cells, which have been shown to play an important role in liver inflammation and other diseases (Figure 4C).
Single-Celled Sequencing Analysis of Gene Expression Profile
To identify the single-cell expression profiles of DNAJB1, CCNL1 and BTG2 in liver I/RI, we analyzed the single-cell dataset GSE171539 and identified a consensus of 13 cell subpopulations, which is similar to the results of previous studies (Figure 5A).19 The expression profile analysis showed that DNAJB1 was mainly expressed in T cells and Kupffer cells, BTG2 was mainly expressed in plasma cells, and CCNL1 was mainly expressed in endothelial cells and plasma cells (Figure 5B and C). These results show the functional diversity of BTG2, CCNL1, and DNAJB1.
Analysis of Co-Expressed Genes
Next, we analyzed the co-expressed genes of DNAJB1, CCNL1, and BTG2. In order to identify potential regulatory mechanisms, we found 240 co-expressed genes of BTG2, 376 co-expressed genes of CCNL1, and 148 co-expressed genes of DNAJB1 (Figure 6A and Tables S5–S7).
 |
Figure 6 Analysis of co-expressed genes. (A) The co-expression genes of BTG2, CCNL1 and DNAJB1 in liver I/RI were analyzed. |
RNA Binding Domain Analysis
Wenhao Jin et al recently used machine learning and deep learning techniques (HydRA) to develop an Artificial Intelligence (AI) prediction of RNA-binding domains that proved to be more than 90% accurate.13 Next, we predicted the RNA-binding domains of DNAJB1, BTG2 and CCNL1 using the HydRA machine learning algorithm, and the results showed that the domain of BTG2 may be located at 64–85aa, the domain of CCNL1 is located at 380–439aa, and the domain of DNAJB1 is located at 188–227aa (Figure 7A and B). The application of RBP2GO database to predict RNA-binding proteins also showed that DNAJB1, CCNL1 and BTG2 were highly likely to be RNA-binding proteins, and it was noteworthy that DNAJB1 and CCNL1 had been captured and identified as RNA-binding proteins by RIC (RNA interactome capture) experiments (Table 1).20
 |
Table 1 RBP2GO Database to Predict RNA-Binding Proteins |
Protein Expression Verification
We then carried out experimental verification. By constructing a mouse liver I/RI model, we conducted immunohistochemistry. Compared with the sham operation group, the expressions of BTG2, CCNL1 and DNAJB1 were significantly up-regulated in the I/RI group (Figure 8A).
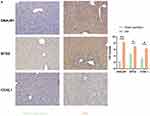 |
Figure 8 Protein expression analysis. (A) Immunohistochemical analysis showed that the expressions of DNAJB1, BTG2 and CCNL1 were up-regulated after liver I/RI. *p<0.05, **p<0.01. |
Discussion
Liver ischemia-reperfusion is a common clinical complication that can easily lead to serious consequences.21 Liver I/RI is an inflammatory disease characterized by massive liver cell damage and death, inflammatory cell infiltration, and explosive cytokine release.22,23 Finding the diagnosis and intervention target of hepatic I/RI can help to detect hepatic I/RI in early clinical stage, formulate treatment plan, and improve the prognosis of patients.
In this study, we focus on 1411 candidate RNA-binding proteins recently discovered by AI machine learning HydRA. Through cross-validation of multiple data sets, we constructed three genetic diagnostic models. Both the internal data set and the external verification set show that the AUC value is more than 0.9, and the DCA curve is good, indicating that the model has good stability and security. It is worth noting that although there was a slight decrease in AUC in the external dataset GSE151648, this may be due to bias in experimental sample selection. We also screened for three key genes (DNAJB1, CCN1 and BTG2). DNAJB1 is a heat shock family protein involved in a wide range of functions such as cell complex assembly and protein folding.24,25 DNAJB1 was found to be involved in both intracellular heat shock response and stress response.24,25 DNAJB1 has also been reported to be involved in promoting liver inflammation.24,25 CCNL1, a cyclin with serine kinase activity, is involved in intracellular cycle regulation and, interestingly, has been reported to be involved in pre-mRNA regulation.26,27 CCNL1 has been reported to be differentially expressed in osteoarthritis and may be involved in osteoarthritis regulation.26,27 BTG2 is an intracellular anti-proliferative protein, which has been reported to form a complex with CCR4-NOT to exert intracellular effects and participate in post-transcriptional regulation of intracellular mRNA.28,29 However, whether BTG2 regulates RNA independently and its binding domain is not fully understood. We used the differentially expressed genes of I/RI to perform functional analysis of GO and KEGG, and the GO analysis showed that I/RI is mainly related to the response to molecular of bacteria origin and cytokine activity pathways. Cytokine activity has been confirmed to be closely related to the development of liver I/RI, and is also the main implementer of the inflammatory response of I/RI.30 KEGG pathway showed that I/RI differential genes were enriched in MAPK, IL-17, NF-KB and other signaling pathways, which have been confirmed to be closely related to inflammation and liver I/RI. Furthermore, we found that the above three genes are mainly enriched in inflammatory pathways such as TNF-α and MAPK, which have been confirmed to play a key role in liver I/RI. These data support the important role of BTG2, DNAJB1 and CCNL1. A large number of studies have also revealed that RNA-binding proteins can participate in the regulation of the above pathway by regulating mRNA metabolism, which also suggests that BTG2, DNAJB1 and CCNL1 may affect the key genes of the above pathway and thus participate in the transmission of the signal pathway.31 Interestingly, our analysis found that BTG2, CCNL1, and DNAJB1 were closely related to mast cell activation. Mast cells were not enriched in the liver under normal circumstances, however, mast cells were involved in liver inflammatory response, and the specific mechanism still needs to be further explored.32 It is worth noting that single-cell sequencing analysis found that DNAJB1, BTG2, and CCNL1 were expressed in large quantities in immune cells, and it was found that gene differential expression in immune cells would lead to phenotypic changes, and then participate in inflammatory immune regulation. It may be a future research direction to study the regulatory mechanism of the above-mentioned three genes in immune cells. By structural analysis of the RNA-binding domains of DNAJB1, CCNL1, and BTG2, we found that the predicted domains by HydRA were mainly distributed in previously unannotated regions, providing a new understanding of the above molecules. In the future, molecular biological techniques such as RNA immunoprecipitation can be used to find the downstream targets of DNAJB1, BTG2, and CCNL1, and explore their correlation with liver I/RI.
Conclusion
In conclusion, in this study, we built a stable diagnostic model, found the key genes of liver I/RI, and conducted systematic functional and structural analysis, providing a new vision for liver I/RI. However, in the future, the effects of the discovered genes (DNAJB1, BTG2, and CCNL1) on signaling pathways (MAPK, JAK-STAT, ERK, JNK, etc.) that play an important role in liver ischemia reperfusion injury can be investigated and their mechanisms of action need to be elucidated.
Data Sharing Statement
All data of the article can be obtained from the corresponding author with reasonable request.
Ethics Statement
This study was conducted in adherence to the guidelines of Good Clinical Practice and the Declaration of Helsinki. The study protocol was reviewed and approved by the Ethics Committee of the Second Hospital of Chongqing Medical University (approval ID:2023-558). All laboratory operations on Animals follow the Guidelines for the Care and Use of Laboratory Animals published by the National Research Council and published by the National Institutes of Health. All mouse procedures were approved by the Bioethics Committee of the Second Hospital of Chongqing Medical University (approval ID:2023-558).
Consent for Publication
All authors agree to publish.
Funding
This project was supported by the Chongqing Natural Science Foundation cstc2020jcyj-msxmX0655.
Disclosure
The authors declare no competing interest in this work.
References
1. Nwaduru C, Baker E, Buff M, et al. Assessing liver viability: insights from mitochondrial bioenergetics in ischemia-reperfusion injury. Transplant Proc. 2024;56(1):228–235. doi:10.1016/j.transproceed.2023.11.019
2. Güven B, Tanoglu A, Ozcelik F, Tanoglu EG, Terzi NK. 4-phenyl butyric acid improves hepatic ischemia/reperfusion and affects gene expression of ABC transporter Abcc5 in rats. Croat Med J. 2023;64(6):391–403. doi:10.3325/cmj.2023.64.391
3. Zhang Y, Qi C, Guo Y, Li X, Zhu Z. Key m6A regulators mediated methylation modification pattern and immune infiltration characterization in hepatic ischemia-reperfusion injury. BMC Med Genomics. 2023;16(1):314. doi:10.1186/s12920-023-01751-0
4. Gao W, Zhang L, Li Z, et al. Nuclear acly protects liver from ischemia-reperfusion injury. Hepatology. 2023. doi:10.1097/HEP.0000000000000692
5. Umman V, Kepil N, Uzun H, Goksoy E. Pre-treatment with pregabalin reduces liver ischemia-reperfusion injury in rats: tissue protection with an analgesic. Eur Rev Med Pharmacol Sci. 2023;27(21):10322–10333. doi:10.26355/eurrev_202311_34307
6. Wu YL, Li TY, Gong XY, et al. Risk factors for myocardial injury during living donor liver transplantation in pediatric patients with biliary atresia. World J Gastrointest Surg. 2023;15(9):2021–2031. doi:10.4240/wjgs.v15.i9.2021
7. Kong E, Zhang Y, Geng X, Zhao Y, Yue W, Feng X. Inhibition of Sirt3 activates the cGAS-STING pathway to aggravate hepatocyte damage in hepatic ischemia-reperfusion injury mice. Int Immunopharmacol. 2024;128:111474. doi:10.1016/j.intimp.2023.111474
8. Völkers M, Preiss T, Hentze MW. RNA-binding proteins in cardiovascular biology and disease: the beat goes on. Nat Rev Cardiol. 2024;21(6):361–378. doi:10.1038/s41569-023-00958-z
9. Avila-Lopez P, Lauberth SM. Exploring new roles for RNA-binding proteins in epigenetic and gene regulation. Curr Opin Genet Dev. 2023;84:102136. doi:10.1016/j.gde.2023.102136
10. Godwin A, Yang WL, Sharma A, et al. Blocking cold-inducible RNA-binding protein protects liver from ischemia-reperfusion injury. Shock. 2015;43(1):24–30. doi:10.1097/SHK.0000000000000251
11. Wen D, Xiao H, Gao Y, Zeng H, Deng J. N6-methyladenosine-modified SENP1, identified by IGF2BP3, is a novel molecular marker in acute myeloid leukemia and aggravates progression by activating AKT signal via de-SUMOylating HDAC2. Mol Cancer. 2024;23(1):116. doi:10.1186/s12943-024-02013-y
12. Wang T, Zhang H. Exploring the roles and molecular mechanisms of RNA binding proteins in the sorting of noncoding RNAs into exosomes during tumor progression. J Adv Res. 2023. doi:10.1016/j.jare.2023.11.029
13. Jin W, Brannan KW, Kapeli K, et al. HydRA: deep-learning models for predicting RNA-binding capacity from protein interaction association context and protein sequence. Mol Cell. 2023;83(14):2595–2611.e11. doi:10.1016/j.molcel.2023.06.019
14. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi:10.1038/s41467-019-09234-6
15. Lu J, Wang X, Feng Z, Chen Y, Wen D, Liu Z. The protective effect of isoflurane pretreatment on liver IRI by suppressing noncanonical pyroptosis of liver macrophages. Int Immunopharmacol. 2021;99:107977. doi:10.1016/j.intimp.2021.107977
16. Kaltenmeier C, Wang R, Popp B, Geller D, Tohme S, Yazdani HO. Role of Immuno-Inflammatory Signals in Liver Ischemia-Reperfusion Injury. Cells. 2022;11(14):2222. doi:10.3390/cells11142222
17. Zhai Y, Busuttil RW, Kupiec-Weglinski JW. Liver ischemia and reperfusion injury: new insights into mechanisms of innate-adaptive immune-mediated tissue inflammation. Am J Transplant. 2011;11(8):1563–1569. doi:10.1111/j.1600-6143.2011.03579.x
18. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi:10.1038/nmeth.3337
19. Wang L, Li J, He S, et al. Resolving the graft ischemia-reperfusion injury during liver transplantation at the single cell resolution. Cell Death Dis. 2021;12(6):589. doi:10.1038/s41419-021-03878-3
20. Caudron-Herger M, Jansen RE, Wassmer E, Diederichs S. RBP2GO: a comprehensive pan-species database on RNA-binding proteins, their interactions and functions. Nucleic Acids Res. 2021;49(D1):D425–D436. doi:10.1093/nar/gkaa1040
21. Wu M, Liu X, Yu Q, Shi J, Guo W, Zhang S. Adelmidrol ameliorates liver ischemia-reperfusion injury through activating Nrf2 signaling pathway. Eur J Pharmacol. 2023;964:176224. doi:10.1016/j.ejphar.2023.176224
22. He Z, Ma C, Yu T, et al. Activation mechanisms and multifaceted effects of mast cells in ischemia reperfusion injury. Exp Cell Res. 2019;376(2):227–235. doi:10.1016/j.yexcr.2019.01.022
23. Klune JR, Tsung A. Molecular biology of liver ischemia/reperfusion injury: established mechanisms and recent advancements. Surg Clin North Am. 2010;90(4):665–677. doi:10.1016/j.suc.2010.04.003
24. Bauer J, Köhler N, Maringer Y, et al. The oncogenic fusion protein DNAJB1-PRKACA can be specifically targeted by peptide-based immunotherapy in fibrolamellar hepatocellular carcinoma. Nat Commun. 2022;13(1):6401. doi:10.1038/s41467-022-33746-3
25. Huang S, Ju W, Zhu Z, et al. Comprehensive and combined omics analysis reveals factors of ischemia-reperfusion injury in liver transplantation. Epigenomics. 2019;11(5):527–542. doi:10.2217/epi-2018-0189
26. Li S, Ma L, Cui R. Identification of novel diagnostic biomarkers and classification patterns for osteoarthritis by analyzing a specific set of genes related to inflammation. Inflammation. 2023;46(6):2193–2208. doi:10.1007/s10753-023-01871-w
27. Ali Khan A, Rodriguez A, Sebert S, et al. The interplay of variants near LEKR and CCNL1 and social stress in relation to birth size. PLoS One. 2012;7(6):e38216. doi:10.1371/journal.pone.0038216
28. Hu Q D, Wang HL, Liu J, et al. Btg2 promotes focal segmental glomerulosclerosis via Smad3-dependent podocyte-mesenchymal transition. Adv Sci. 2023;10(32):e2304360. doi:10.1002/advs.202304360
29. Ameerul A, Almasmoum H, Pavanello L, Dominguez C, Winkler GS. Structural Model of the human BTG2-PABPC1 complex by combining mutagenesis, NMR chemical shift perturbation data and molecular docking. J Mol Biol. 2022;434(14):167662. doi:10.1016/j.jmb.2022.167662
30. Lv J, Zhu X, Xing C, et al. Stimulator of interferon genes (STING): key therapeutic targets in ischemia/reperfusion injury. Biomed Pharmacother. 2023;167:115458. doi:10.1016/j.biopha.2023.115458
31. Chai RC, Chang YZ, Chang X, et al. YTHDF2 facilitates UBXN1 mRNA decay by recognizing METTL3-mediated m6A modification to activate NF-κB and promote the malignant progression of glioma. J Hematol Oncol. 2021;14(1):109. doi:10.1186/s13045-021-01124-z
32. Bernard JK, Marakovits C, Smith LG, Francis H. Mast cell and innate immune cell communication in cholestatic liver disease. Semin Liver Dis. 2023;43(2):226–233. doi:10.1055/a-2104-9034
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.







