Back to Journals » Journal of Inflammation Research » Volume 17
Discovery and Validation of Ferroptosis-Associated Genes of Ulcerative Colitis
Authors Zhu J , Wu Y, Ge X, Chen X, Mei Q
Received 27 March 2024
Accepted for publication 3 July 2024
Published 9 July 2024 Volume 2024:17 Pages 4467—4482
DOI https://doi.org/10.2147/JIR.S463042
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Jiejie Zhu,1,2 Yumei Wu,1,2 Xiaoyuan Ge,1,2 Xinwen Chen,1,2 Qiao Mei1,2
1Department of Gastroenterology, The First Affiliated Hospital of Anhui Medical University, Hefei City, Anhui Province, People’s Republic of China; 2Key Laboratory of Digestive Diseases of Anhui Province, Hefei, People’s Republic of China
Correspondence: Qiao Mei, The First Affiliated Hospital of Anhui Medical University, 218 Jixi Road, Hefei, Anhui Province, 230022, People’s Republic of China, Email [email protected]
Background: Ulcerative colitis (UC) is a long-lasting idiopathic condition, but its precise mechanisms remain unclear. Meanwhile, evidence has demonstrated that ferroptosis seems to interlock with the progress of UC. This research sought to identify hub genes of UC related to ferroptosis.
Methods: First, the relevant profiles for this article were obtained from GEO database. From the FerrDb, 479 genes linked to ferroptosis were retrieved. Using analysis of the difference and WGCNA on colonic samples from GSE73661, the remaining six hub genes linked to ferroptosis and UC were discovered. Through logistic regression analyses, the diagnostic model was constructed and was then evaluated by external validation using dataset GSE92415. Afterwards, the correlation between immune cell filtration in UC and hub genes was examined. Finally, a mice model of colitis was established, and the results were verified using qRT-PCR.
Results: We acquired six hub genes linked to ferroptosis and UC. In order to create a diagnostic model for UC, we used logistic regression analysis to screen three of the six ferroptosis related genes (HIF1A, SLC7A11, and LPIN1). The ROC curve showed that the three hub genes had outstanding potential for disease diagnosis (AUC = 0.976), which was subsequently validated in samples from GSE92415 (AUC = 0.962) and blood samples from GSE3365 (AUC = 0.847) and GSE94648 (AUC = 0.769). These genes might be crucial for UC immunity based upon the results on the immune system. Furthermore, mouse samples examined using qRT-PCR also verified our findings.
Conclusion: In conclusion, the findings have important implications for ferroptosis and UC, and these hub genes may also offer fresh perspectives on the aetiology and therapeutic approaches of UC.
Keywords: ulcerative colitis, ferroptosis, hub genes, immune, bioinformatics analysis
Introduction
Inflammatory bowel disease (IBD) is a kind of chronic disease that encompasses ulcerative colitis (UC) and Crohn’s disease. The incidence of IBD, especially UC, is increasing rapidly globally, and how to diagnose and manage UC is a major public health problem.1 Due to intestinal inflammation that results in diarrhea, abdominal pain, and rectal bleeding, UC lowers the quality of people’s lives.2 The pathogenesis of UC relates to genetic factors, gut microbiota dysbiosis, immune response disorders and leukocyte recruitment.3 Additionally, oxidative stress is a potential etiological for UC.4 Intestinal inflammation is accompanied by excessive production of reactive oxygen and nitrogen metabolites, which in turn results in DNA damage, apoptosis, and accelerating inflammation.5,6 The presence of excessive reactive oxygen metabolites and the imbalance of antioxidants were found in UC tissues compared with normal tissues.7,8 Studies in DSS-induced murine models have demonstrated that regulation of redox signaling pathways, such as NF-κB signaling pathway and the nuclear factor-erythroid 2-related factor 2 (Nrf2) pathway, can alleviate inflammation symptoms.9,10 The protracted and erratic clinical course is characterized by remission and aggravating episodes that alternate.11 Despite aggressive medication and interventions, a small percentage of UC patients present with a rapidly progressing type of colitis commonly known as fulminant illness.12
Several forms of cell death, including autophagy and apoptosis, come into play under physiological and pathological conditions.13 Ferroptosis is a kind of iron-dependent cell death, which has recently emerged and can induce the formation of reactive species (ROS), is the key to inflammation and human diseases.14 Ferroptosis is strongly connected to the pathologic and physiologic processes of numerous diseases, including carcinogenesis, nervous system disorders, renal damage, and hematological systemic diseases, according to recent studies.15–18 In addition, there is a possible relationship between ferroptosis and UC. Recently, a population-based case–control trial with parts of Japanese patients on risk factors showed an association between a higher dietary intake of iron and an increased risk of UC.19 Xu M et al first reported the mechanism and significance of ferroptosis in the intestinal epithelial cell death of UC.20 Curculigoside is a natural ingredient with antioxidative and anti-inflammatory properties that could shield UC patients from ferroptosis via upregulating the generation and expression of GPX4.21 Furthermore, inhibitors of ferroptosis including ferrostatin-1 (Fer-1) have been validated in models of UC, which could effectively ameliorate clinical symptoms and pathological phenotypes of DSS-induced colitis as well as TNBS-induced colitis.22,23 Overall, these results imply a key role for ferroptosis in UC.
In recent years, gene microarrays and integrated bioinformatics analysis offer fresh perspectives on the etiology of various diseases. This work found six hub genes linked to ferroptosis and UC from the GEO database by using a range of bioinformatics methods. Figure 1 displays the study’s procedure.
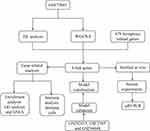 |
Figure 1 The process workflow for this study. |
Material and Methods
Obtaining Data and Difference Analyzing
The NCBI Gene Expression Omnibus public database (GEO) provided the gene expression data needed in this research. Datasets that met the following filtering criteria were used for this study: ① expression profiling by array; ② Homo sapiens; ③ dataset containing more than ten UC samples and healthy controls. The raw data of GSE73661, which was annotated by GPL6244, contains information on RNA levels. One hundred and sixty-six colonic biopsies of UC and 12 control samples in GSE73661 were collected. The validation data set GSE92415 annotated by GPL13158 included 162 UC samples and 21 controls. Samples from two datasets GSE3365 and GSE94648 were extracted from peripheral blood. GSE3365 annotated by GPL96 included 26 UC samples and 42 control samples, and GSE94648 annotated by GPL19109 included 25 UC samples and 22 control samples. After removing irrelevant header information, a large number of gene expression data from UC and control patient biopsies of intestinal tissue were analyzed and compared with the aid of the “limma” package in R program. Some genes in compliance with the filter criteria including p_adj < 0.05 and abs(logFC) > 2 were screened as DEGs. The R packages “ggplot2” and “pheatmap” were employed to generate heat maps and volcano plots of DEGs. In addition, supplementary box 1 lists 479 genes associated with ferroptosis that were downloaded from the FerrDb website and used for this study’s investigation.
Weighted Correlation Network Analysis (WGCNA)
This is a common procedure to build the gene co-expression network via “WGCNA” package in R software.24 Firstly, we utilized gene-expression arrays to calculate each gene’s MAD (Median Absolute Deviation) and then eliminated any samples that were aberrant. To create a similarity matrix, the correlation coefficient between every gene pair was determined. Subsequently, the adjacency was converted into a topological overlap matrix (TOM), which was then used to calculate the corresponding dissimilarity (1-TOM) and quantify the network connectivity of a gene, which was defined as the sum of its adjacency with all other genes for network Gene ration. A minimum size of 30 was required for the Genes dendrogram in order to perform average linkage hierarchical clustering based on the TOM-based dissimilarity measure for the purpose of categorizing Genes with comparable expression profiles into Gene modules. The key modules connected to ferroptosis were identified using the module eigengene (ME) and the module membership (MM). We determined the dissimilarity of the characteristic genes of the modules, selected a cut line for the module dendrogram, and combined several modules in order to further investigate the modules. We also combined modules that were closer together than 0.25. Ultimately, 21 co-expression modules were obtained; nevertheless, a gray module was obtained, which indicated a group of genes that could not be allocated to any of those modules.
Discovery of Hub Genes
The VennDiagram tool in the R program was applied to extract the intersection of genes from DEGs, elements derived from WGCNA, and genes relevant to ferroptosis in order to acquire hub genes connected with ferroptosis and UC. Violin plots were used to illustrate the hub gene differential expression in UC and control. The t-test and Mann–Whitney U-test were the hypothesis tests employed. The former was applied if the data had a normal distribution; in the absence of one, the latter.
Functional Enrichment Evaluation
Functional enrichment analysis was performed to investigate the physiological mechanisms of hub genes influencing ulcerative colitis. Firstly, we examined the biological processes of gene ontology that these genes were implicated in. The concluded findings were displayed as a chord diagram using the R program’s “GOplot” utility. Gene Set Enrichment Analysis (GSEA) was then used to identify each gene’s unique function. The GSEA software, version 3.0, was acquired from the GSEA website. According to the median gene expression level, the samples were split into two groups: high expression and low expression. Biological Signatures Database (http://www.gsea-msigdb.org/gsea/downloads.jsp) was used to download the c2.cp.kegg.v7.4.symbols.gmt subset in order to assess the relevant signal cascades and cellular processes based on gene expression patterns. We have 5 as the minimum and 5000 as the maximum gene set values. The value of p < 0.05 and FDR < 0.25 possess the statistical sense.
Building and Testing the Logistic Regression Model
First, the logistic regression model was built to help differentiate UC patients from controls effectively. Then, receiver operating characteristic (ROC) curve analyses were performed using the pROC package of R to determine the logistic regression model’s diagnostic and predictive value. In general, the closer the ROC curve is to the upper left corner, the better the diagnostic performance is represented. Finally, the classification efficacy was evaluated by external validation using dataset GSE92415.
Exploration of Immune Infiltration
In order to examine the constitution of immunizing and defending system, the proportion of 22 different kinds of human immune cells in the UC and controls was obtained utilizing the CIBERSORT algorithm, with p < 0.05 considered to be significant differences between the two groups.25 In addition, Spearman correlation analysis was conducted to assess correlations on immune cells in microenvironment using the R tool.
Establishment of the Mouse Model of Colitis
Anhui Medical University’s Laboratory Animals Ethics Committee gave its clearance for all procedures involving animals in this experiment. For seven days, 3% dextran sulfate sodium (DSS) (MP Biomedicals, with an average molecular weight of 36000–50000) dissolved in sterile distilled water was given ad libitum to male C57BL/6 WT mice (6–8 weeks old) in order to induce colitis. Throughout the trial, mice in the control group were given regular access to pure water. There were six mice in each group. During the experiment period, survival and clinical parameters, such as body weight, rectal bleeding and stool consistency of mice were monitored daily. DAI grading standards refer to Kihara et al.26 Mice were killed by cervical dislocation, and the entire colon in each mouse was removed from caecum to anus. Colon lengths were measured using a ruler. The section of colon tissues used for qRT-PCR was stored at −80°C.
Histological Analysis
At autopsy, the colon tissue from model mice was flushed with ice saline and excised. Each mouse’s tissue specimens were preserved for one day using neutral formaldehyde solution and then processed for paraffin embedding and sectioning. The histopathologic change in mucosal structure was observed by hematoxylin and eosin staining under a light microscope. Histological scores were determined based on four individual inflammatory parameters as reported previously: crypt damage, inflammation severity, inflammation extent, and percent involvement.26
2.9 qRT-PCR
With the help of the TRIzol reSuperMix agent (Invitrogen), all RNA molecules were obtained from the murine colon tissues. Using HiScript III RT and ChamQ SYBR qPCR Master Mix (Vazyme) in accordance with protocols, the procedure of reverse transcription and amplifying a piece of DNA were carried out under specific thermal cycling conditions. After normalizing each gene’s relative mRNA level to GAPDH, the final results were presented as 2−ΔΔCt. The primers used in this paper are listed as follows: Forward 5’-3’: GGTTGTCTCCTGCGACTTCA, reverse 5’-3’: TGGTCCAGGGTTTCTTACTCC for GAPDH. Forward 5’-3’: GGTTTTCAGTGTCACCACGC, reverse 5’-3’: CAAGGGTGGGCAAAATGTGG for LPIN1. Forward 5’-3’: GAATGAAGTGCACCCTAACAAG, reverse 5’-3’: GAGGAATGGGTTCACAAATCAG for HIF1A. Forward 5’-3’: TCTGGTCTGCCTGTGGAGTA, reverse 5’-3’: CAAAGGACCAAAGACCTCCA for SLC7A11. Forward 5’-3’: ACTGTGCCACTCGTCCTGAT, reverse 5’-3’: TCTCCAGATTGTCCAAGGCT for PARP9. Forward 5’-3′: TTACGTCCATCGTGGACAGC, reverse 5′-3′: TGGGCTGGGTGTTAGTCT TA, for NOS2. Forward 5′-3′: -CCCACACTGAGCACGGACGG, reverse 5′-3′: TTGCGGGGCAGCACCTTTCG for IDO1.
Statistics
R software or the statistical program GraphPad Prism were used for all the statistical computations. After checking the data for homogeneous distribution, a t-test for equal variances, otherwise known as the Mann–Whitney U-test was chosen to see the difference between two groups.
Results
Discovery of Hub Genes Related to UC and Ferroptosis
Using the screening criterion “p_adj < 0.05 and abs(logFC) > 2”, we first extracted 898 differentially expressed genes from GSE73661 in order to find genes associated with UC (Supplementary Table 1). A volcano diagram was used to display these DEGs (Figure 2A). The top 20 genes with differential expression were shown on a heatmap (Figure 2B). A weighted gene co-expression network was built using the expression profiles of 2,3303 genes and 178 samples that were obtained from GSE73661 after the genes had been filtered and aberrant samples eliminated. In Figure 3A and B, the mean connected value was 146.40 on a scale independence of 0.76 considering the soft threshold power was adjusted to 6. Through the use of dynamic tree cutting, twenty-one distinct modules co-expressed were procured on the premise of a minimum module size of thirty and a cut height of 0.25 (Figure 3C). Following that, assessments of every single module’s connection with clinical characteristics were carried out. As shown in Figure 3D, the dark green module exhibiting the strongest and positive relationship has a profound impact on UC (r = 0.50, p = 2.0e−12), whereas the cyan module represented the highest negative correlation with UC and was also keenly relevant (r = −0.66, p = 1.8e−23). Afterwards, we got a gene set consisting of 479 genes from FerrDb and compared them to the 898 DEGs and 90 dark green module genes, six hub genes were found, but no genes associated with ferroptosis and UC were obtained by taking the intersections of 898 DEGs, 479 ferroptosis-associated genes and 87 cyan module genes (Figures 3 and 4). Therefore, the dark green module, containing 90 genes with the biggest related coefficient, could further be used in the next step. Moreover, module membership and gene importance were shown to be substantially connected, according to a correlation analysis, with these genes corresponding to both phenotype and module (cor = 0.57, p = 1.5e−49; Figures 3 and 4). These six genes have the key function in regulating various biological processes. Specifically, they participate in regulation of cell death pathway, the regulation of stress responses, autophagy. Among the iron metabolic issues, these genes are responsible for maintaining iron ion homeostasis, responding to iron ion fluctuations, and heme binding (Figures 3 and 4). Violin plots demonstrated that all six genes were expressed in UC as well as controls by analyzing the massive medical information from GSE73661, with higher expression level seen in UC (Figure 5). Furthermore, Supplementary Figure 1 displays the expression of these genes in colon tissue samples from GSE92415. In addition, the expression of these genes in blood samples from GSE3365 and GSE94648 was shown in Supplementary figures 2 and 3.
 |
Figure 5 The expression of hub genes in the UC and control groups of colonic mucosa samples. (A) HIF1A, (B) SLC7A11, (C) LPIN1, (D) IDO1, (E) NOS2, (F) PARP9. |
Enriched Biological Processes and Pathways
Enrichment studies were leveraged to gain insights into the possible biological activities of these genes. GSEA in Figure 6 has revealed that these genes exhibit significant associations with gut-related biological pathways, encompassing the intestinal immune network responsible for IgA production and membrane structures of partial intestinal mucosa epithelium tight junction. Additionally, the analysis has demonstrated their involvement in inflammation-related signaling cascades, including cell apoptosis, activation and release of inflammatory cells and mediators such as chemotactic factors, T cellular receptor and NOD-like receptor.
 |
Figure 6 GSEA illustrated the enriched pathways of the hub genes. (A) HIF1A, (B) SLC7A11, (C) LPIN1, (D) IDO1, (E) NOS2, (F) PARP9. |
A Diagnostic Model Applicable to UC
Through constructing logistic regression, HIF1A, SLC7A11, LPIN1 were selected, which could effectively differentiate UC patients from controls, boasting an AUC value of 0.976 (Figure 7A). Subsequently, the model underwent rigorous validation in samples from another dataset. In the GSE92415 dataset, the AUCs of the model reached 0.962, further corroborating its reliability (Figures 7B). The model was also validated in blood samples. The AUCs of the models in GSE3365 and GSE94648 were 0.847 and 0.769, respectively (Figure 7C and D). Colon samples tended to be more representative of UC pathology than blood samples, which may explain the better diagnostic performance of the former. However, blood samples are easier to obtain by noninvasive tests instead of by endoscope. In summary, the positive outcomes obtained from colon tissue samples and blood samples revealed that this model may hold significant guiding value when it comes to the identification of UC patients in clinical practice.
Immune Infiltration and Immune Cell Correlation Analysis
Gene changes to the microenvironment are highly influential in determining the diagnosis of disease and drug sensitivity. The study employed the CIBERSORT algorithm to assess the fraction of immune cells in 166 UC samples and 12 control samples. The resulting histogram (Figure 8A) illustrates this process. A boxplot was utilized to compare the immune cell infiltration of the control and UC samples (Figure 8B). In terms of cell ratio, the UC group had noticeably greater proportions of dendritic cells activated (P=4.9e-5), naive B cells (p = 6.5e-3), CD4 memory resting T cells (p = 5.8e-5), follicular helper T cells (P=1.2e-3), M0 macrophages (P=1.1e-5), M1 macrophages (P=1.8e-4), Mast cells activated (P=1.9e-5), neutrophils (P=1.9e-3), and lower proportions of CD8 T cells (P=6.5e-5), regulatory T cells (Tregs) (P=6.1e-6), NK cells activated (P=7.8e-9), M2 macrophages (P=6.0e-4), mast cells resting (P=6.3e-7) compared with the control group. In the following step, an analysis of the immune cells’ relationships was conducted (Figure 8C). These findings raised the possibility that hub genes are significant components of maintaining immune homeostasis and transition to disease.
Confirmation of the Hub Genes in the Mice Used as Model for Colitis
Ultimately, as shown in Figure 9A, we validated the gene expression patterns of hub genes in a murine model of DSS-induced acute colitis, thereby ensuring the precision and reliability of our findings. There are significant weight loss and colon shortening in DSS-induced colitis, indicating a successful model was established (Figure 9B–D). Furthermore, the disease activity index (DAI) of the DSS group was notably worse compared to the control group (Figure 9C). After hematoxylin-eosin staining, the light microscope capacitates colon tissue from all the mice to be observed carefully. There was a breakdown of the intestinal mucosa’s typical structure and the crypt cells died away in the model group (Figure 9E). The histological score of the DSS group exhibited an obviously higher level compared to that of the other group (Figure 9F). Several studies have found that ferroptosis was elevated in mice with colitis brought on by DSS.27,28 The level of six genes was subsequently tested by qRT-PCR assay. Our Results demonstrated that there was a substantial difference in the relative mRNA amounts of HIF1A, SLC7A11, LPIN1, IDO1, and NOS2 in the DSS group compared to the control group (Figure 10A–E). Intriguingly, PARP9 expression had no significant association with feeding DSS to mice (Figure 10F).
Discussion
Ferroptosis is caused by the overwhelming peroxidation of lipids, which was found to be involved in the progression of numerous diseases, especially intestinal diseases. Some drugs and molecular targets exert anti-colitis and anti-carcinogenesis effects via inhibiting ferroptosis, such as sorafenib, maternal embryonic leucine zipper kinase (MELK).29,30 Inhibiting ferroptosis can lessen intestinal injury in non-infectious inflammatory conditions like intestinal I/R injury and IBD, while inducing ferroptosis with pharmacological activators can inhibit the migration, invasion, and proliferation of colorectal neoplasms, suggesting a dual role for ferroptosis in different intestinal diseases.31–34 Although there are several ways of assaying ferroptosis from multiple aspects, there is no general agreement on a standard criteria for defining the occurrence of ferroptosis. To elucidate ferroptosis in vivo, it needs to identify specific ferroptosis indicators and a better alternative. Therefore, more research is imperative to delve deeper into ferroptosis in intestinal diseases.
Chronic inflammation in patients with UC exhibits a close association with ferroptosis. Six genes that bridge the connection between UC and ferroptosis were identified through a meticulous analysis of data derived from one dataset GSE73661 publicly available. A quantitative analysis of GO revealed that these genes are intricately implicated in the regulation of stress response, cell death, and the acquisition and metabolic process of iron. Several findings have revealed that stress could trigger dysfunction of the gut barrier, alteration of intestinal dysbiosis, harm to self-renewal of stem cells in the intestines, ultimately leading to inflammation and cell damage.35–37 Autophagy is essential to the preservation of epithelial homeostasis during stress.38 In addition, Lonicerin alleviated UC by interrupting the inflammasome pathway mediated by autophagy.39 Dioscin effectively promoted autophagy in the DSS-induced mouse model by targeting the AMPK-mTOR signaling pathway.40 Interestingly, 3-methyladenine could pharmacologically suppress autophagy, which also affected the induction of ferroptosis by ionizing radiation,41 indicating that autophagic machinery may contribute to the occurrence of ferroptosis under certain conditions. Herein, GSEA in Figure 6 further corroborated that these hub genes were intimately correlated with processes connected to the gut and inflammation-related signaling pathways.
Three out of the six genes, namely HIF1A, SLC7A11, and LPIN1, were chosen for the formation of a diagnostic model that could potentially aid in the clinical diagnosis of UC. HIF1A, a component of the hypoxia-inducible factor (HIF) family, possesses the ability to bind to the promoters of numerous target genes, thereby initiating the transcription of a diverse array of genes that are involved in metabolic processes in cells, adaptability to hypoxia, and the regulation of immune responses.42 There is a significant correlation between hypoxia and inflammation. IBD is distinguished by inflammation susceptible to hypoxia, coinciding with inflammation and hypoxia at sites of inflammation.43 Ferroptosis is involved in osteoclasts, and it has been demonstrated that HIF-1α reduces ferritinophagy by inhibiting the development of autophagosomes, thereby shielding osteoclasts from ferroptosis.44 Additionally, in acute kidney injury, ferroptosis is regulated by the activation of HIF and further promotes mitophagy and other multiple potential mechanisms.45 SLC7A11, commonly referred to as xCT, efficiently facilitates the uptake of cystine, which would inhibit ferroptosis and oxidative stress.46 Recent investigations have unequivocally confirmed that SLC7A11 is overexpressed in a diverse range of tumors and promotes growth and drug resistance of tumors partly through mediating ferroptosis.47,48 Tu H et al revealed that ferroptosis, apoptosis, and autophagy-dependent cell death are all influenced by SLC7A11.49 LPIN1 belongs to the family of lipin proteins and is necessary for maintaining normal lipid homeostasis.50 Ferroptosis is relevant to oxidative degradation of lipids. Studies have found that lipid metabolism abnormality affect ferroptosis and governs the sensitivity to ferroptosis via regulating phospholipid peroxidation, alongside other cellular processes tied to it.51,52 Lipid metabolic genes including ACSL4 and LPCAT3 have been reported to facilitate ferroptosis.50,53 As a transcriptional regulator of lipid metabolism, LPIN1 performs crucial functions within the nucleus and membranes of different organelles, including endoplasmic reticulum membrane, endolysosomes and mitochondrial membranes.54 Mice lacking LPIN1 have an accumulation of abnormal mitochondria as a result of an autophagic flux blockage.55 However, whether LPIN1 is involved in ferroptosis during the development of UC is still unclear, which would be an interesting direction.
Herein, we delved deeper into the large number of inflamed cells in the illness and normal group. The findings demonstrated a large rise in M0/M1 macrophages and much decreased M2 macrophages in UC. Gut macrophages are thought to involve the clinical manifestations and disease processes of UC.56 Research revealed that as compared to the epithelium tissues of the normal group, the intestinal mucosal macrophages in the colonic tissue of individuals with active UC were in abundance and likely to be polarized into classical activated pro-inflammatory phenotype (M1), which were consistent with our study.57,58 Hence, reestablishing the M1/M2 macrophage polarization balance may be a new class of targets for UC. In patients with IBD, including UC, activation of dendritic cells (DCs) results in up-regulated expression of microbial recognition receptors on DCs and further produces elevated levels of cytokines that are pathologically relevant.59 The increased abundance of neutrophils, the induction of neutrophil extracellular traps, and several associated proteins has been reported in UC patients, confirming that neutrophils play a pivotal part in the pathophysiological process of UC.60,61 Furthermore, our study observed a decrease in Tregs, which are a subset of T cells in charge of regulating tissue inflammation and maintaining immune homeostasis. However, findings on the intestinal and circulating Tregs in IBD patients remain controversial. In contrast to healthy controls and inactive IBD patients, the ratio of circulating Tregs was much lower in active IBD patients, although the proportion of intestinal Tregs was found to be larger in damaged tissues compared to regions without inflammation.62 It has been shown that ferroptosis is implicated with the lethal accumulation of reactive oxygen species (ROS), leading to the increased production of inflammatory mediators and enhancing immune cell infiltration, with implications for aberrant inflammation in UC.63 It is interesting to note that interleukin-6 (IL-6), a marker of M1 macrophages, could cause lipid peroxidation and disrupt cell iron homeostasis, which in turn causes ferroptosis.64 In addition, it was discovered that ferroptosis is associated with the formation of neutrophil extracellular traps.65 These findings imply that immune cells are also involved in the regulation of ferroptosis. The crosstalk between them provides a new inspiration for the treatment of UC and deserves further exploration.
Mice were used to develop the DSS-induced UC model successfully in order to support the findings of the bioinformatics analysis. There is a combination of these reasons for the use of acute DSS model in this study. Firstly, chronic colitis model is more often used in adaptive immunity and complications such as tissue fibrosis and carcinogenesis, while acute colitis models may be more suitable for oxidative damage and acute stress research.66 Recent studies have revealed that ferroptosis was induced significantly in colonic tissues from acute DSS-treated mice.67 In addition, ulcerative colitis is a long-term chronic disease, but it is often accompanied by acute exacerbation in the clinical course, such as bleeding ulcers and erosions in active UC patients.68 Moreover, we will continue to explore these genes in chronic colitis models in the future. A dramatic upregulation of SLC7A11, HIF1A, LPIN1, IDO1, and NOS2 expression was seen in the UC model, as evidenced by qRT-PCR. However, the lower mRNA pression of PARP9 in the colitis model detected by qRT-PCR did not agree with the previous finding based on the bioinformatics analysis. The result may flow from several causes. Firstly, it is important to note that our bioinformatics results were based on human colonic mucosa, whereas the qRT-PCR assay was conducted on mouse colon tissue specimens. Consequently, there exists a fundamental heterogeneity between the two experimental platforms. These results need to be investigated further for human colon mucosa in subsequent experiments, not only at the gene level but also at the protein level. Second, the UC patients were treated with vedolizumab therapy or infliximab therapy, which may have an influence on colonic mucosal gene expression. In addition, the sample size was small. Notably, existing researches have shown that dysregulated glutathione production and cysteine absorption are caused by the inhibition and degradation of SLC7A11, which also triggers ferroptosis.69–71 Indeed, we found that the expression of SLC7A11 both in human UC tissues and DSS-induced mice model was up-regulated. These results and underlying mechanisms need to be intensively investigated further. Furthermore, although there are a large number of regulators that directly or indirectly affect iron accumulation and lipid peroxidation to regulate ferroptosis, there are still many unresolved questions, such as what is the physiological and pathological function of ferroptosis in human health and disease including UC, and how ferroptosis affects the prognosis of UC, and what are the specific immune signals associated with ferroptosis at different stages of inflammation. Further research is still needed to investigate the complicated machinery and regulation of ferroptosis in UC.
Conclusion
In summary, we successfully obtained six hub genes intricately linked to ferroptosis and UC through the application of bioinformatics approaches, namely HIF1A, SLC7A11, LPIN1, IDO1, NOS2, PARP9. We delved into the biological processes and pathways in which these genes are engaged, thereby gaining valuable insights into the pathogenesis of ulcerative colitis. Utilizing logistic regression analysis, we developed a diagnostic model capable of accurately identifying patients suffering from ulcerative colitis. Furthermore, these hub genes have been discovered to be interconnected with diverse immune cells, hinting at their potential significance in shaping the immune microenvironment. Nonetheless, additional research is imperative to unveil the precise roles these genes play in UC.
Data Sharing Statement
The data generated and thoroughly analyzed throughout this study are available for retrieval from the corresponding author upon receipt of a reasonable request.
Acknowledgments
We express our gratitude to the team at the Gene Expression Omnibus (GEO) for allowing us to utilize their invaluable data.
Ethics Approval and Consent to Participate
Ethical approval for this study was granted by the Animal Ethical Committee of Anhui Medical University (Approval No.20221095). The animal experiments were in line with the Guidelines for the Care and Use of Laboratory Animals of the National Academy of Sciences. The human sample data involved in this study conformed to national and institutional ethical guidelines and were approved by the Ethics Committee of the First Affiliated Hospital of Anhui Medical University (PJ2022-07-27).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Barnes EL, Loftus EV, Kappelman MD. Effects of race and ethnicity on diagnosis and management of inflammatory bowel diseases. Gastroenterology. 2021;160(3):677–689. doi:10.1053/j.gastro.2020.08.064
2. Gros B, Kaplan GG. Ulcerative colitis in adults: A review. JAMA. 2023;330(10):951–965. doi:10.1001/jama.2023.15389
3. Ordás I, Eckmann L, Talamini M, Baumgart DC, Sandborn WJ. Ulcerative colitis. Lancet. 2012;380(9853):1606–1619. doi:10.1016/S0140-6736(12)60150-0
4. Zhou M, Xu W, Wang J, et al. Boosting mTOR-dependent autophagy via upstream TLR4-MyD88-MAPK signalling and downstream NF-κB pathway quenches intestinal inflammation and oxidative stress injury. EBioMedicine. 2018;35:345–360. doi:10.1016/j.ebiom.2018.08.035
5. Kang R, Li R, Dai P, Li Z, Li Y, Li C. Deoxynivalenol induced apoptosis and inflammation of IPEC-J2 cells by promoting ROS production. Environ Pollut. 2019;251:689–698. doi:10.1016/j.envpol.2019.05.026
6. Sahoo DK, Heilmann RM, Paital B, et al. Oxidative stress, hormones, and effects of natural antioxidants on intestinal inflammation in inflammatory bowel disease. Front Endocrin. 2023;14:1217165. doi:10.3389/fendo.2023.1217165
7. Kruidenier L, Kuiper I, Lamers CB, Verspaget HW. Intestinal oxidative damage in inflammatory bowel disease: semi-quantification, localization, and association with mucosal antioxidants. J Pathol. 2003;201(1):28–36. doi:10.1002/path.1409
8. Cao W, Vrees MD, Kirber MT, Fiocchi C, Pricolo VE. Hydrogen peroxide contributes to motor dysfunction in ulcerative colitis. Am J Physiol Gastrointest Liver Physiol. 2004;286(5):G833–G843. doi:10.1152/ajpgi.00414.2003
9. Yuan SN, Wang MX, Han JL, et al. Improved colonic inflammation by nervonic acid via inhibition of NF-κB signaling pathway of DSS-induced colitis mice. Phytomedicine. 2023;112:154702. doi:10.1016/j.phymed.2023.154702
10. Guo H, Guo H, Xie Y, et al. Mo3Se4 nanoparticle with ROS scavenging and multi-enzyme activity for the treatment of DSS-induced colitis in mice. Redox Biol. 2022;56:102441. doi:10.1016/j.redox.2022.102441
11. Feuerstein JD, Cheifetz AS. Ulcerative colitis: epidemiology, diagnosis, and management. Mayo Clin Proc. 2014;89(11):1553–1563. doi:10.1016/j.mayocp.2014.07.002
12. Rubin DT, Ananthakrishnan AN, Siegel CA, Sauer BG, Long MD. ACG Clinical Guideline: ulcerative Colitis in Adults.. Am J Gastroenterol. 2019;114(3):384–413. doi:10.14309/ajg.0000000000000152
13. Kist M, Vucic D. Cell death pathways: intricate connections and disease implications. EMBO J. 2021;40(5):e106700. doi:10.15252/embj.2020106700
14. Liang D, Minikes AM, Jiang X. Ferroptosis at the intersection of lipid metabolism and cellular signaling. Mol Cell. 2022;82(12):2215–2227. doi:10.1016/j.molcel.2022.03.022
15. Sun X, Ou Z, Chen R, et al. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology. 2016;63(1):173–184. doi:10.1002/hep.28251
16. Lane DJR, Ayton S, Bush AI. Iron and Alzheimer’s Disease: An update on emerging mechanisms. J Alzheimers Dis. 2018;64(s1):S379–S395.
17. Martin-Sanchez D, Ruiz-Andres O, Poveda J, et al. Ferroptosis, but not necroptosis, is important in nephrotoxic folic Acid-Induced AKI. J Am Soc Nephrol. 2017;28(1):218–229. doi:10.1681/ASN.2015121376
18. Li J, Cao F, Yin HL, et al. Ferroptosis: past, present and future. Cell Death Dis. 2020;11(2):88. doi:10.1038/s41419-020-2298-2
19. Kobayashi Y, Ohfuji S, Kondo K, et al. Association between dietary iron and zinc intake and development of ulcerative colitis: a case-control study in Japan. J Gastroenterol Hepatol. 2019;34(10):1703–1710. doi:10.1111/jgh.14642
20. Xu M, Tao J, Yang Y, et al. Ferroptosis involves in intestinal epithelial cell death in ulcerative colitis. Cell Death Dis. 2020;11(2):86. doi:10.1038/s41419-020-2299-1
21. Wang S, Liu W, Wang J, Bai X. Curculigoside inhibits ferroptosis in ulcerative colitis through the induction of GPX4. Life Sci. 2020;259:118356. doi:10.1016/j.lfs.2020.118356
22. Chen Y, Zhang P, Chen W, Chen G. Ferroptosis mediated DSS-induced ulcerative colitis associated with Nrf2/HO-1 signaling pathway. Immunol Lett. 2020;225:9–15. doi:10.1016/j.imlet.2020.06.005
23. Xu J, Liu S, Cui Z, et al. Ferrostatin-1 alleviated TNBS induced colitis via the inhibition of ferroptosis. Biochem Biophys Res Commun. 2021;573:48–54. doi:10.1016/j.bbrc.2021.08.018
24. Gu X, Lai D, Liu S, et al. Hub genes, diagnostic model, and predicted drugs related to iron metabolism in Alzheimer’s disease. Front Aging Neurosci. 2022;14:949083. doi:10.3389/fnagi.2022.949083
25. Zhao S, Chi H, Yang Q, et al. Identification and validation of neurotrophic factor-related gene signatures in glioblastoma and Parkinson’s disease. Front Immunol. 2023;14:1090040. doi:10.3389/fimmu.2023.1090040
26. Kihara N, de la Fuente SG, Fujino K, Takahashi T, Pappas TN, Mantyh CR. Vanilloid receptor-1 containing primary sensory neurones mediate dextran sulphate sodium induced colitis in rats. Gut. 2003;52(5):713–719. doi:10.1136/gut.52.5.713
27. Huang F, Zhang S, Li X, Huang Y, He S, Luo L. STAT3-mediated ferroptosis is involved in ulcerative colitis. Free Radic Biol Med. 2022;188:375–385. doi:10.1016/j.freeradbiomed.2022.06.242
28. Zhang X, Li W, Ma Y, et al. High-fat diet aggravates colitis-associated carcinogenesis by evading ferroptosis in the ER stress-mediated pathway. Free Radic Biol Med. 2021;177:156–166. doi:10.1016/j.freeradbiomed.2021.10.022
29. Yuan S, Wei C, Liu G, et al. Sorafenib attenuates liver fibrosis by triggering hepatic stellate cell ferroptosis via HIF-1α/SLC7A11 pathway. Cell Prolif. 2022;55(1):e13158. doi:10.1111/cpr.13158
30. Tang B, Zhu J, Fang S, et al. Pharmacological inhibition of MELK restricts ferroptosis and the inflammatory response in colitis and colitis-propelled carcinogenesis. Free Radic Biol Med. 2021;172:312–329. doi:10.1016/j.freeradbiomed.2021.06.012
31. Chen X, Li J, Kang R, Klionsky DJ, Tang D. Ferroptosis: machinery and regulation. Autophagy. 2021;17(9):2054–2081. doi:10.1080/15548627.2020.1810918
32. Long D, Mao C, Huang Y, Xu Y, Zhu Y. Ferroptosis in ulcerative colitis: potential mechanisms and promising therapeutic targets. Biomed Pharmacoth. 2024;175:116722. doi:10.1016/j.biopha.2024.116722
33. Li Y, Feng D, Wang Z, et al. Ischemia-induced ACSL4 activation contributes to ferroptosis-mediated tissue injury in intestinal ischemia/reperfusion. Cell Death Differ. 2019;26(11):2284–2299. doi:10.1038/s41418-019-0299-4
34. Li D, Wang Y, Dong C, et al. CST1 inhibits ferroptosis and promotes gastric cancer metastasis by regulating GPX4 protein stability via OTUB1. Oncogene. 2023;42(2):83–98. doi:10.1038/s41388-022-02537-x
35. Weiss GA, Hennet T. Mechanisms and consequences of intestinal dysbiosis. Cell Mol Life Sci. 2017;74(16):2959–2977. doi:10.1007/s00018-017-2509-x
36. Guo J, Lou X, Gong W, et al. The effects of different stress on intestinal mucosal barrier and intestinal microecology were discussed based on three typical animal models. Front Cell Infect Microbiol. 2022;12:953474. doi:10.3389/fcimb.2022.953474
37. Hageman JH, Heinz MC, Kretzschmar K, van der Vaart J, Clevers H, Snippert HJG. Intestinal regeneration: Regulation by the Microenvironment. Dev Cell. 2020;54(4):435–446. doi:10.1016/j.devcel.2020.07.009
38. Foerster EG, Mukherjee T, Cabral-Fernandes L, Rocha JDB, Girardin SE, Philpott DJ. How autophagy controls the intestinal epithelial barrier. Autophagy. 2022;18(1):86–103. doi:10.1080/15548627.2021.1909406
39. Lv Q, Xing Y, Liu J, et al. Lonicerin targets EZH2 to alleviate ulcerative colitis by autophagy-mediated NLRP3 inflammasome inactivation. Acta Pharm Sin B. 2021;11(9):2880–2899. doi:10.1016/j.apsb.2021.03.011
40. Li H, Pang B, Nie B, et al. Dioscin promotes autophagy by regulating the AMPK-mTOR pathway in ulcerative colitis. Immunopharm Immuno. 2022;44(2):238–246. doi:10.1080/08923973.2022.2037632
41. Zhou H, Zhou YL, Mao JA, et al. NCOA4-mediated ferritinophagy is involved in ionizing radiation-induced ferroptosis of intestinal epithelial cells. Redox Biol. 2022;55:102413. doi:10.1016/j.redox.2022.102413
42. Corcoran SE, O’Neill LA. HIF1α and metabolic reprogramming in inflammation. J Clin Invest. 2016;126(10):3699–3707. doi:10.1172/JCI84431
43. Xue X, Ramakrishnan S, Anderson E, et al. Endothelial PAS domain protein 1 activates the inflammatory response in the intestinal epithelium to promote colitis in mice. Gastroenterology. 2013;145(4):831–841. doi:10.1053/j.gastro.2013.07.010
44. Ni S, Yuan Y, Qian Z, et al. Hypoxia inhibits RANKL-induced ferritinophagy and protects osteoclasts from ferroptosis. Free Radic Biol Med. 2021;169:271–282. doi:10.1016/j.freeradbiomed.2021.04.027
45. Li W, Xiang Z, Xing Y, Li S, Shi S. Mitochondria bridge HIF signaling and ferroptosis blockage in acute kidney injury. Cell Death Dis. 2022;13(4):308. doi:10.1038/s41419-022-04770-4
46. Koppula P, Zhuang L, Gan B. Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell. 2021;12(8):599–620. doi:10.1007/s13238-020-00789-5
47. Liu L, Liu R, Liu Y, et al. Cystine-glutamate antiporter xCT as a therapeutic target for cancer. Cell Biochem Funct. 2021;39(2):174–179. doi:10.1002/cbf.3581
48. Okuno S, Sato H, Kuriyama-Matsumura K, et al. Role of cystine transport in intracellular glutathione level and cisplatin resistance in human ovarian cancer cell lines. Br J Cancer. 2003;88(6):951–956. doi:10.1038/sj.bjc.6600786
49. Tu H, Tang LJ, Luo XJ, Ai KL, Peng J. Insights into the novel function of system Xc- in regulated cell death. Eur Rev Med Pharmacol Sci. 2021;25(3):1650–1662. doi:10.26355/eurrev_202102_24876
50. Reue K. The lipin family: mutations and metabolism. Curr Opin Lipidol. 2009;20(3):165–170. doi:10.1097/MOL.0b013e32832adee5
51. Chen X, Kang R, Kroemer G, Tang D. Broadening horizons: the role of ferroptosis in cancer. Nat Rev Clin Oncol. 2021;18(5):280–296. doi:10.1038/s41571-020-00462-0
52. Pope LE, Dixon SJ. Regulation of ferroptosis by lipid metabolism. Trends Cell Biol. 2023;33(12):1077–1087. doi:10.1016/j.tcb.2023.05.003
53. Dixon SJ, Winter GE, Musavi LS, et al. Human haploid cell genetics reveals roles for lipid metabolism genes in nonapoptotic cell death. ACS Chem Biol. 2015;10(7):1604–1609. doi:10.1021/acschembio.5b00245
54. Zhang P, Reue K. Lipin proteins and glycerolipid metabolism: roles at the ER membrane and beyond. Biochim Biophys Acta Biomembr. 2017;1859(9):1583–1595. doi:10.1016/j.bbamem.2017.04.007
55. Zhang P, Verity MA, Reue K. Lipin-1 regulates autophagy clearance and intersects with statin drug effects in skeletal muscle. Cell Metab. 2014;20(2):267–279. doi:10.1016/j.cmet.2014.05.003
56. Na YR, Stakenborg M, Seok SH, Matteoli G. Macrophages in intestinal inflammation and resolution: a potential therapeutic target in IBD. Nat Rev Gastroenterol Hepatol. 2019;16(9):531–543. doi:10.1038/s41575-019-0172-4
57. Lissner D, Schumann M, Batra A, et al. Monocyte and M1 Macrophage-induced barrier defect contributes to chronic intestinal inflammation in IBD. Inflamm Bowel Dis. 2015;21(6):1297–1305. doi:10.1097/MIB.0000000000000384
58. Zhang M, Li X, Zhang Q, Yang J, Liu G. Roles of macrophages on ulcerative colitis and colitis-associated colorectal cancer. Front Immunol. 2023;14:1103617. doi:10.3389/fimmu.2023.1103617
59. Hart AL, Al-Hassi HO, Rigby RJ, et al. Characteristics of intestinal dendritic cells in inflammatory bowel diseases. Gastroenterology. 2005;129(1):50–65. doi:10.1053/j.gastro.2005.05.013
60. Hansberry DR, Shah K, Agarwal P, Agarwal N. Fecal myeloperoxidase as a biomarker for inflammatory bowel disease. Cureus. 2017;9(1):e1004. doi:10.7759/cureus.1004
61. Bennike TB, Carlsen TG, Ellingsen T, et al. Neutrophil extracellular traps in ulcerative colitis: a proteome analysis of intestinal biopsies. Inflamm Bowel Dis. 2015;21(9):2052–2067. doi:10.1097/MIB.0000000000000460
62. Duan S, Cao Y, Chen P, Yang Y, Zhang Y. Circulating and intestinal regulatory T cells in inflammatory bowel disease: A systemic review and meta-analysis. Int Rev Immunol. 2024. doi:10.1080/08830185.2023.2249525
63. Ocansey DKW, Yuan J, Wei Z, Mao F, Zhang Z. Role of ferroptosis in the pathogenesis and as a therapeutic target of inflammatory bowel disease (Review). Int J Mol Med. 2023;51(6):53. doi:10.3892/ijmm.2023.5256
64. Yang D, Liang H, Zhu X, et al. Farnesoid X Receptor Protects Murine Lung against IL-6-promoted Ferroptosis Induced by Polyriboinosinic-Polyribocytidylic Acid. Am J Respir Cell Mol Biol. 2024;70(5):364–378. doi:10.1165/rcmb.2023-0172OC
65. Lv T, Xiong X, Yan W, Liu M, Xu H, He Q. Mitochondrial general control of amino acid synthesis 5 like 1 promotes nonalcoholic steatohepatitis development through ferroptosis-induced formation of neutrophil extracellular traps. Clin Transl Med. 2023;13(7):e1325. doi:10.1002/ctm2.1325
66. Wirtz S, Popp V, Kindermann M, et al. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat Protoc. 2017;12(7):1295–1309. doi:10.1038/nprot.2017.044
67. Wu Y, Ran L, Yang Y, et al. Deferasirox alleviates DSS-induced ulcerative colitis in mice by inhibiting ferroptosis and improving intestinal microbiota. Life Sci. 2023;314:121312. doi:10.1016/j.lfs.2022.121312
68. Leppkes M, Lindemann A, Gößwein S, et al. Neutrophils prevent rectal bleeding in ulcerative colitis by peptidyl-arginine deiminase-4-dependent immunothrombosis. Gut. 2022;71(12):2414–2429. doi:10.1136/gutjnl-2021-324725
69. Yang WS, Stockwell BR. Ferroptosis: death by Lipid Peroxidation. Trends Cell Biol. 2016;26(3):165–176. doi:10.1016/j.tcb.2015.10.014
70. Liu GZ, Xu XW, Tao SH, Gao MJ, Hou ZH. HBx facilitates ferroptosis in acute liver failure via EZH2 mediated SLC7A11 suppression. J Biomed Sci. 2021;28(1):67. doi:10.1186/s12929-021-00762-2
71. Chen Q, Zheng W, Guan J, et al. SOCS2-enhanced ubiquitination of SLC7A11 promotes ferroptosis and radiosensitization in hepatocellular carcinoma. Cell Death Differ. 2023;30(1):137–151. doi:10.1038/s41418-022-01051-7
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.








