Back to Journals » Journal of Asthma and Allergy » Volume 18
Genetic Variant rs1801275 in Atopic Dermatitis: Prevalence and Clinical Implications in Vietnamese Population
Authors Nguyen THN, Le DHH , Huynh TTM, Le TT, Nguyen TH , Cao Dinh H , Chau TV, Nguyen HM
Received 8 April 2025
Accepted for publication 10 June 2025
Published 30 June 2025 Volume 2025:18 Pages 1065—1078
DOI https://doi.org/10.2147/JAA.S528514
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Luis Garcia-Marcos
Tuan Huu Ngoc Nguyen,1,2 Duong Hoang Huy Le,1,2 Thi Thi Mai Huynh,3 Thoi Thi Le,4 Thinh Hung Nguyen,1,2 Hung Cao Dinh,5,6 Tro Van Chau,7 Ha Minh Nguyen1,2,8
1Biomedical Research Center, Pham Ngoc Thach University of Medicine, Ho Chi Minh, Vietnam; 2Medical Biochemistry & Molecular Biology Department, Fundamental Sciences and Basic Medical Sciences, Pham Ngoc Thach University of Medicine, Ho Chi Minh, Vietnam; 3Outpatient Department, Hospital of Dermato-Venereology, Ho Chi Minh City, Vietnam; 4Department of Medical Laboratory Science, Faculty of Nursing and Medical Technology, University of Medicine and Pharmacy at Ho Chi Minh City, Ho Chi Minh City, Vietnam; 5Department of Internal Medicine, Faculty of Medicine, Pham Ngoc Thach University of Medicine, Ho Chi Minh City, Vietnam; 6Department of Internal Medicine, Faculty of Medicine, Nguyen Tat Thanh University, Ho Chi Minh City, Vietnam; 7Department of Dermatology of Pham Ngoc Thach University of Medicine, Faculty of Medicine, Ho Chi Minh, Vietnam; 8Laboratory Department, Nguyen Tri Phuong Hospital, Ho Chi Minh, Vietnam
Correspondence: Ha Minh Nguyen, Email [email protected]
Background: Atopic dermatitis (AD), a chronic inflammatory skin condition, affects up to 20% of children and 10% of adults globally, driven by a type 2 immune response via IL-4 and IL-13 through IL-4Rα. The rs1801275 variant in IL-4Rα gene, a glutamine-to-arginine substitution (Q576R), increases AD severity and atopic comorbidities. This study examines rs1801275’s prevalence and clinical impact in Vietnamese population.
Methods: A cross-sectional study (January–May 2021) with 113 AD patients (Hanifin and Rajka criteria) and 213 healthy controls has been conducted. Demographics, clinical features, and SCORAD severity were assessed via questionnaires and dermatologist evaluations. rs1801275 variant was genotyped using allele-specific real-time PCR. Frequencies were compared, and associations with AD severity were analyzed using Fisher’s Exact Test, Kruskal–Wallis test, and logistic regression, adjusting for age and sex.
Results: Allele frequencies (A: 82.74% vs 79.58%; G: 17.26% vs 20.42%) and genotypes of AD patient and control groups, respectively, showed no significant difference (p = 0.315), indicating no link to AD susceptibility. However, the G allele was associated with higher SCORAD severity in the dominant model (AG+GG vs AA: median 40 vs 30.5, p = 0.010; OR 4.67, p = 0.005) and additive model (p = 0.023), with a dose effect (AA: 30.5, AG: 39, GG: 49.65). Age group independently predicted severity (OR 2.31– 2.43, p < 0.05).
Conclusion: The rs1801275 variant correlates with increased severity in G allele carriers, per SCORAD, in dominant model. These findings support personalized AD management in Vietnam, though larger studies are needed for GG genotypes.
Keywords: atopic dermatitis, IL-4Rα, rs1801275, Q576R, genetic variant, SCORAD severity, Vietnamese population
Introduction
Atopic dermatitis (AD), a chronic inflammatory skin condition, affects millions of people worldwide, with prevalence rates reaching 20% in children and 10% in adults.1 In Vietnam, local studies have observed that the proportion of AD is 15.3% in infants under the age of five and 6.7% in adults.2,3 The disease presents a significant public health challenge due to its debilitating symptoms, impact on quality of life, and association with various comorbid conditions, such as asthma and allergic rhinitis.4
The pathology of AD is complex as an interaction consequence of genetic and environmental factors leading to the alterations of skin barrier and operation of the immune system.5 The Th2 immune pathway is a critical component of the adaptive immune response that plays a central role in allergic inflammation, which is also essential in all development phases of AD. The Th2 cells produce cytokines such as IL-4, IL-5, and IL-13, which drive the production of IgE antibodies and the differentiation of eosinophils, key players in allergic reactions. The IL-4Rα chain is a common component of both the IL-4 and IL-13 receptors, making it a pivotal molecule in Th2 signaling.6
As a matter of fact, genome-wide association studies (GWAS) have identified multiple genetic loci that are associated to AD, including the rs1801275 (A→G) variant (Q576R).7 The Q576 variant, located in the cytoplasmic domain of the IL-4Rα chain, is critical for signal transduction and regulation. Studies have shown that the R576 variant enhances IL-4 signaling, leading to Th2 cytokine response amplification, and even promotes the conversion of induced Treg (iTreg) cells towards a T helper 17 (TH17) cell fate.8,9 In mouse model, following epicutaneous sensitization with either ovalbumin or house dust mite, the IL-4RαR576/R576 and IL-4RαQ576/R576 mice displayed skin inflammation significantly more severe with the high expression of IL-4 and IL13 and antigen-specific IgE antibody levels than the IL-4RαQ576/Q576 controls.10
The augmented prevalence of AD in Asian populations around the world, notably in urban areas, urges for the population-specific understanding of disease susceptibility and development.11 This need is further stressed by the Asian particular genetic background as well as the introduction of IL-4Rα-targeted medications, such as dupilumab, requiring investigations on efficacy predictors, including IL-4Rα genotypes, for optimal AD treatment outcomes.12
This study aims to bridge these knowledge gaps in Vietnamese population by investigating the prevalence and clinical implications of the IL-4Rα rs1801275 variant. Using a comparative cross-sectional design, we compare genotype and allele frequencies between AD patients and healthy controls, while assessing associations of the rs1801275 variant with disease stage, SCORAD severity, and atopic comorbidities. Our findings contribute to genetic database of Southeast Asia and facilitate the precision medicine practice in AD management in Vietnam, potentially leading to patient outcome improvement.
Patients, Materials and Methods
Patients and Study Design
A comparative cross-sectional study was conducted on Vietnamese patients with atopic dermatitis (AD), diagnosed per Hanifin and Rajka diagnostic criteria, recruited consecutively from the outpatient clinic of Ho Chi Minh City Dermatology Hospital between January and May 2021.13 Patients with systemic autoimmune diseases, other chronic dermatoses, or recent treatment (within 4 weeks prior to enrollment) with immunosuppressive agents, systemic or topical corticosteroids, or regular use of moisturizers were excluded to minimize potential confounding effects on disease severity. The control group comprised healthy Vietnamese volunteers from Pham Ngoc Thach University of Medicine, screened via structured interviews and clinical examination by a trained physician to exclude personal or family history of atopic diseases. Both the patient and control groups were predominantly residents of Ho Chi Minh City. With the G allele frequency in the general East Asian population reported as 0.178, sample size was calculated to detect a 15% allele frequency difference with 80% power and α = 0.05, requiring at least 106 cases and 212 controls.14 Informed consent was obtained from all participants, with parental consent for children under 15 and assent from older minors. At the end of the study, we enrolled 113 AD patients and 213 control individuals. Data of all participants is eligible for further analysis and report.
Patients’ Data Collection
Data was collected from each patient using a standardized, pre-tested questionnaire administered by trained research staff to ensure consistency and minimize interviewer bias. The questionnaire captured epidemiological characteristics (age, sex, residential address, nationality, education level, occupation, marital status), personal and family medical history (including atopic dermatitis, asthma, and allergic rhinitis), age of disease onset, factors triggering exacerbations (classified as environmental [eg, dust mites, pollen], dietary [eg, seafood, dairy], or psychological [eg, stress], based on patient recall over the preceding 6 months), and subjective symptoms (intensity of pruritus assessed via a 10-point visual analog scale (VAS), ranging from 0 [no itch] to 10 [severe itch]). Dividing patients into <2, 2–12, and >12 years acknowledges distinct age-related variations in disease presentation, etiology, and treatment approaches.15 Clinical examinations were performed by two board-certified dermatologists trained in the Hanifin and Rajka diagnostic criteria (1980) and SCORAD (Scoring Atopic Dermatitis) index.16 Disease severity was categorized as mild (<25), moderate (25–50), or severe (>50) based on SCORAD scores, and disease staging (acute, subacute, chronic) was determined using the European Academy of Dermatology and Venereology (EADV) guidelines. Photographic documentation of skin lesions was obtained at a fixed distance of 50 cm under 500 lux lighting to validate clinical findings and support severity assessments. All data were recorded in a secure case report form, with double-entry verification performed to ensure accuracy.
rs1801275 Genotyping
Venous blood samples (3 mL) were collected in EDTA-anticoagulated tubes, stored at 2–8°C, and transported to the Biomedical Research Center at Pham Ngoc Thach University of Medicine within 4 hours for DNA extraction. Genomic DNA was extracted from leukocytes using the TopPure Blood Extraction Kit (ABT, Vietnam) following the manufacturer’s silica column-based protocol within 6 hours after collection to minimize degradation. DNA purity was assessed using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA), with an OD260/OD280 ratio of 1.7–2.0 considered acceptable. Extracted DNA was stored at −20°C until genotyping. For genotyping, DNA was diluted to 1.25 ng/µL in TE buffer for downstream PCR amplification. The rs1801275 variant in the IL-4Rα gene was genotyped using allele-specific PCR with the SensiFAST SYBR Lo-ROX Kit (Bioline, UK) on a Quant Studio 5 Real-Time PCR System (ABI, ThermoFisher Scientific, USA).17 Two PCRs with 3 primers IL-4Rα-AF (5′-CCCCACCAGTGGCTATCA-3′), IL-4Rα-GF (5′-CCCCACCAGTGGCTATCG-3′) and IL-4Rα-R (common primer) (5′-GCCTTGTAACCAGCCTCTCC-3′) targeting allele A (PCR-A) and allele G (PCR-G) separately were conducted in parallel. Each 10 µL PCR reaction contained SensiFAST SYBR Lo-ROX Master Mix 2X, forward primer (400 nM), reverse primer (400 nM), 2.5 ng of template DNA in TE buffer. Amplification conditions included an initial denaturation at 95°C for 2 minutes, followed by 35 cycles of denaturation at 95°C for 5 seconds and annealing/extension with fluorescence capture at 70°C for 15 seconds. Each run included a no-template control (NTC) and a known heterozygous positive control to detect contamination and confirm assay specificity, respectively. The genotype of a given sample was determined based on the cycle threshold (CT) of both PCRs. The AA or GG genotype will have the CTPCR-A or CTPCR-G lower than the other at least 3 cycles, whereas GA genotype will have the difference between CTPCR-A and CTPCR-G less than 2 cycles. All samples were processed in duplicate, and discrepant results were resolved by a third amplification. Residual DNA samples were stored at −20°C in a secure biobank, as detailed in the Ethical Approval section.
Statistical Analysis
Data analysis was performed using SPSS version 20.0 (SPSS Inc., Chicago, IL, USA) and R software version 4.2.1 (R Foundation for Statistical Computing). Genotype and allele frequencies were calculated for both case and control groups and assessed for Hardy-Weinberg equilibrium (HWE) using the Chi-square test, with p > 0.05 indicating equilibrium. Categorical variables (eg, genotype distribution, disease severity categories) were presented as frequencies and percentages, while continuous variables (eg, SCORAD scores) were tested for normality using the Shapiro–Wilk test. Normally distributed variables were reported as means ± standard deviations (SD), and non-normally distributed variables as medians with interquartile ranges (IQR). Differences in allele and genotype frequencies between cases and controls were evaluated using the Fisher Exact Test when expected cell counts were <5, and the Chi-square test otherwise. For SCORAD scores, differences between allele groups (A vs G) were assessed using the Mann–Whitney U-test, and between genotype groups (AA vs AG vs GG) using the Kruskal–Wallis test, followed by post-hoc Dunn’s test with Bonferroni correction for multiple comparisons. To explore associations between the rs1801275 variant and clinical characteristics (eg, SCORAD severity, age of onset), logistic regression models were employed, adjusting for potential confounders including age, sex, and family history of atopy; odds ratios (ORs) with 95% confidence intervals (CIs) were reported. Missing data (<5% of total entries) were handled using listwise deletion, and sensitivity analyses were performed to assess the robustness of findings. Data visualizations were generated using Flourish Studio (https://flourish.studio/) and Biorender (https://www.biorender.com/).
Ethical Approval
This study was approved by the Ethical Committee of Pham Ngoc Thach University of Medicine under two decisions: Number 440/TĐHYKPNT-HĐĐĐ, dated December 11, 2020, and Number 771/HĐĐĐ-ĐHYD, dated October 24, 2022. The research was conducted in accordance with the principles of the Declaration of Helsinki and the International Conference on Harmonisation-Good Clinical Practice (ICH-GCP) guidelines. Written informed consent was obtained from all participants prior to enrollment. For participants under 15 years of age, parental or legal guardian consent was secured, while older minors (15–18 years) provided additional assent. All personal data were anonymized using unique identification codes, and electronic records were stored in a password-protected database accessible only to authorized researchers. Biological samples were labeled with corresponding codes and preserved at −20°C in a secure biobank for a maximum of 5 years post-study, after which they will be destroyed per institutional policy. Participants were informed of their right to withdraw from the study at any time without consequence. No financial incentives were provided, ensuring voluntary participation.
Results
Demographic Data and Clinical Characteristics
A comprehensive overview of the characteristics of the study population, including the patient group (n =113) and the control group (n = 213), is presented in Table 1. The patient group exhibited a wide age range (7 months to 87 years, median 11 years, IQR 6–29), reflecting AD’s broad demographic impact, with a notable proportion of early-onset cases (<2 years: 30.09%). The median age was significantly lower in the patient group compared to the control group (37, IQR 30–43), p < 0.001, Mann–Whitney U-test. Gender distribution was relatively balanced in patients (55.75% female) and controls (62.9% female) (p = 0.208). Clinically, AD patients showed a mix of disease stages (acute and subacute: 38.93% each; chronic: 22.12%) and severity levels by SCORAD (mild: 30.09%; moderate: 53.10%; severe: 16.81%). All patients reported itching (100%), and most exhibited typical morphology (73.45%), chronic or recurrent dermatitis (93.81%), and dry skin (99.12%). Atopic comorbidities were present, with 12.39% reporting asthma and 15.93% allergic rhinitis, alongside a strong family history of atopic diseases (eg, AD in 50.45%).
 |
Table 1 Epidemiological and Clinical Characteristics of the Study Populations |
IL-4Rα rs1801275 Frequencies
The rs1801275 variant’s allele and genotype frequencies between AD patients and controls are illustrated and compared in Table 2. The A allele predominated in both groups (patients: 82.74%; controls: 79.58%), with the G allele less frequent (patients: 17.26%; controls: 20.42%), showing no significant difference (p = 0.315, Fisher’s Exact Test).
 |
Table 2 Genotypes and Allelic Frequencies of the IL-4Rα rs1801275 Variant in the Study Population |
The Hardy-Weinberg equilibrium test was performed separately on the patient group, the control group, and the combined population. When compared to the expected populations, the distribution of allele A and allele G was in equilibrium, with no significant differences observed across all three groups, yielding HWE p-values of 0.835, 0.556, and 0.812, respectively (Figure 1).
This chart visually represents the frequency and distribution patterns of alleles and genotypes of the IL-4Rα rs1801275 variant between the observed population groups (in red lines) and the expected population groups (in orange-yellow lines), in three population: (A) patient group; (B) control group; and (C) overall population (patients + controls). When comparing each pair (Chi-square test), the area enclosed within the colored lines shows a nearly overlapping shape across all axes, suggesting that the surveyed population pairs achieve HWE (observed frequencies closely match expected frequencies) (all pHWE > 0.05).
Association of the IL-4Rα rs1801275 Variant with AD Susceptibility
To clarify the impact of the variant genotypes on the disease susceptibility to AD, we examined the differences in genotype frequencies across various genetic models between the control and patient groups (Table 2). In the additive model, genotype frequencies were AA (67.25% vs 65.26%), AG (30.97% vs 28.64%), and GG (1.78% vs 6.10%) for patients and controls, respectively, with p = 0.177. Similarly, the recessive model (AA vs AG+GG) and the overdominant model (AA+AG vs GG) showed no difference. The dominant model (AA+AG vs GG) suggested a trend toward lower GG prevalence in patients (1.78%) than controls (6.10%), though not significant (p = 0.097). The Hardy-Weinberg equilibrium test was performed separately on the patient group, the control group, and the combined population. When compared to the expected populations, the distribution of allele A and allele G was in equilibrium, with no significant differences observed across all three groups, yielding HWE p-values of 0.835, 0.556, and 0.812, respectively (Figure 1).
This chart visually represents the frequency and distribution patterns of alleles and genotypes of the IL-4Rα rs1801275 variant between the observed population groups (in red lines) and the expected population groups (in orange-yellow lines), in three population: (A) patient group; (B) control group; and (C) overall population (patients + controls). When comparing each pair (Chi-square test), the area enclosed within the colored lines shows a nearly overlapping shape across all axes, suggesting that the surveyed population pairs achieve HWE (observed frequencies closely match expected frequencies) (all pHWE > 0.05).
Association of the IL-4Rα rs1801275 Variant with AD Phenotypes
When examining the association of rs1801275 with AD phenotypes across genetic models, no significant associations were found with gender, age group, personal or family atopic history, or disease stage (p > 0.05, data not shown). However, SCORAD severity showed notable findings in the additive, dominant and overdominant model, for “Mild vs Moderate-Severe” and “Mild vs Moderate vs Severe” (Table 3).
 |
Table 3 Association of the IL-4Rα rs1801275 Variant with the Severity by SCORAD Across Genetic Models |
To provide a deeper insight into the differences in disease severity across genotypes, we analyzed quantitative SCORAD scores across four genetic models. The results are presented in Figure 2 (median and IQR quantitative SCORAD scores are not shown). This figure illustrates the differences in quantitative SCORAD scores among genotype groups across the four genetic models, providing visual evidence supporting the use of genotype (particularly AG+GG) as a predictive marker for AD severity.
In the additive model, the median SCORAD score progressively increased from AA (30.5) → AG (39) → GG (49.65), suggesting a dose effect of the G allele, where more G alleles correspond to more severe AD. The IQR also followed an increasing trend (20.5–41 → 29–47 → 40.3–59); however, the IQR for GG was very broad and based on a small sample size (n = 2), reducing reliability. The p-value 0.023 indicated statistically significant differences among the three groups. The Kruskal–Wallis test (non-parametric) was appropriate for comparing three groups with non-normally distributed data. Thus, the additive model confirms the trend of increasing AD severity with a higher number of G alleles (AA < AG < GG), with substantial statistical evidence.
In the dominant model (AA vs AG+GG), the median SCORAD score for AG+GG (40) was significantly higher than that for AA (30.5), indicating that individuals carrying at least one G allele had a higher severity of AD. The IQR of AG+GG (30.9–47) was higher and had minimal overlap with the IQR of AA (20.5–41), reinforcing the difference in SCORAD distribution between the two groups (p = 0.010). Thus, the dominant model suggests that the AA genotype is associated with milder AD (lower median SCORAD), while AG+GG is linked to more severe AD (p = 0.010). The G allele exhibits a dominant effect, increasing disease severity.
In the recessive model (AA+AG vs GG), the median SCORAD score for GG (49.65) was much higher than that for AA+AG (33), suggesting that GG is associated with more severe AD. However, the IQR of GG (40.3–59) was extremely broad and based on a very small sample size (n = 2), reducing the reliability of the result. The IQR of AA+AG (22–43) had minimal overlap with GG, but the difference was not statistically significant (p = 0.154), due to the small GG sample size, which reduced the statistical power of the test.
In the overdominant model (AA+GG vs AG), the median SCORAD score for AG (39) was higher than that for AA+GG (30.75), indicating that the AG heterozygous genotype is associated with more severe AD compared to the AA or GG homozygous genotypes. The IQR of AG (29–47) was higher and had minimal overlap with the IQR of AA+GG (21–41), supporting the distinction between the two groups (p = 0.028).
Predictive Model of AD Severity
When considering the contribution of related factors to the severity of AD, we develop a predictive model to classify disease severity into mild and moderate-severe groups based on SCORAD, using independent variables such as genotype, gender, age group, personal history, family history, and age of onset. This prediction model (Multivariable Logistic Regression Model) was performed using the genotype variable of the IL-4Rα rs1801275 variant for four genetic models: additive, dominant, recessive and overdominant. The results are presented in Figure 3, which clearly visualizes the role of related factors, making it easier to identify key risk factors. They indicate that the IL-4Rα rs1801275 variant (G allele) and age group are the two main predictors of moderate-to-severe atopic dermatitis compared to mild cases. Other factors (gender, personal or family history, age of onset) do not significantly contribute to predicting disease severity (OR ≈ 1, p > 0.05).
Multivariate logistic regression analysis is used to have adjusted OR, representing the risk of moderate-to-severe disease compared to mild disease was adjusted after exclusion of non-significant variables. The results from Table 4 show that, consistently across all genetic models examined, age group is a contributing independent factor to the increased severity of AD, with OR ranging from 2.31 to 2.41 and 95% CI not including the value 0. This means that each unit increase in age group increased the odds by 2.31–2.41 times, with p < 0.05. This aligns with the clinical reality that AD may become more severe with age due to cumulative damage or immune changes. Older age is a factor to consider in patient management, independent of genotype. When considering genotype characteristics, in the recessive model, the GG genotype (n = 2) was exclusively found in the moderate-to-severe SCORAD group, making regression analysis infeasible for this model. In the dominant model (AA vs AG+GG), the AG+GG genotype showed the highest risk increase, with an odds ratio of 4.67 and strong statistical significance (p = 0.005), indicating that the G allele plays a major role in increasing the risk of moderate-to-severe disease, even when carrying only one allele (AG). This is the most compelling model. In the additive model, the risk increased 4.52 times (p = 0.005), demonstrating a dosage effect of the G allele, with a gradual risk increase from AA → AG → GG. However, due to the limited GG data, the distinction between AG and GG could not be clearly established. In the overdominant model, the AG genotype increased the risk 4.03 times (p = 0.011), suggesting that the heterozygous genotype may be particularly significant. The significant association between genotypes and moderate-to-severe AD directly addresses the study’s aim to link rs1801275 with clinical severity, supporting its role in disease phenotype rather than susceptibility.
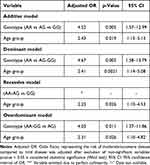 |
Table 4 Adjusted Odds Ratios for Predictors of Mild vs Moderate-to-Severe Disease Severity |
Thus, genotypes containing the G allele (especially AG+GG) serve as important predictors of AD severity, supporting risk stratification and personalized treatment. The dominant model is the most useful tool for risk prediction in clinical practice due to its clarity and strong statistical significance. Increasing the sample size in future studies, particularly for the GG genotype group, will help confirm the effects in the recessive and additive models. Additionally, providing information on age group classification and other potential confounding factors (such as gender and disease duration) will further strengthen the validity of these conclusions.
To conduct sensitivity analysis, we performed logistic regression employing a bootstrap resampling method with one thousand iterations, each involving a sample size of one hundred. ORs and their corresponding 95% CIs were determined for the genotypes of dominant model (AA vs AG-GG) and age group within the mild and moderate-to-severe SCORAD subgroups. The results showed that, for the dominant model, the mean OR was 6.77 (95% CI: 6.38–7.16), while the median OR was 4.93 (IQR: 3.30–7.72). Similarly, for the age group, the mean OR was 2.84 (95% CI: 2.72–2.97), with a median OR of 2.42 (IQR: 1.81–3.31). These findings reinforce the robustness of the associations observed in our primary analysis.
Discussion
Atopic dermatitis, as part of the “atopic march”, is one of the most common skin problems, affecting up to 20% of children around the globe.1 AD is the consequence of an unfavorable interaction between genetic and environmental factors.18
The underlying mechanisms of AD are largely based in immune dysregulation while highlighting the central role of the immune system dysregulation, characterized by a skewing towards a T-helper cell type 2 (Th2) immune response.19 The Th2-dominant state is featured by the augmented production of specific cytokines, including interleukin-4 (IL-4), interleukin-5 (IL-5), interleukin-13 (IL-13), and interleukin-31 (IL-31) 3, which drive the inflammatory processes and contribute to characteristic manifestations of atopic diseases while involving both B and T cells.20
IL-4Rα Q576R, result of the nucleotide substitution at position 1727, is associated with increased expression of the IL-4Rα on lymphocytes and increased STAT6 phosphorylation, leading to exaggerated Type-2 inflammation.9 Several studies have investigated the association of the IL-4Rα Q576R polymorphism with the risk and severity of atopic diseases, but data related to atopic dermatitis in particular is relatively limited. The rs1801275 (Q576R) polymorphism has been associated with atopic dermatitis susceptibility and severity, exacerbation of allergic skin inflammation and family history of atopy.10,21–23 One important observation is that this variant seems not to be associated with IgE-normal atopic diseases.24
Our study is the first investigation of the IL-4Rα rs1801275 variant in Vietnamese atopic dermatitis (AD) patients, revealing its limited role in disease susceptibility but a significant association with severity. It is noteworthy that data analysis was conducted with rigorous methodology, including Hardy-Weinberg equilibrium confirmation (p = 0.835 patients, 0.556 controls, 0.812 combined, Figure 1), ensuring no genetic bias in the population. Age adjustment in logistic regression (Figure 3) mitigated the significant age difference between patients and controls (p = <0.001, Table 1), while bootstrap sensitivity analysis reinforced result reliability, notably in the dominant model (p = 0.010).
We observed no notable difference in G allele frequency between AD patients (17.26%) and controls (20.42%, p = 0.315), suggesting that rs1801275 is not a primary risk factor for AD onset in this population. The G allele frequency in Vietnamese population (17.26–20.42%) aligns with estimates for Asian populations (~20%) but is markedly lower than in Caucasians (~35%).14,25 The rarity of GG in our patient group (1.78% vs 6.10% in controls) may explain the absence of a similar association, potentially reflecting population-specific genetic structures, or epistasis. Understanding epistasis is crucial for unraveling the complex genetic architecture of multifactorial diseases like atopy, where the risk of developing the condition is influenced by the combined effects of multiple genes and environmental exposures. Within the IL-4Rα gene, the Q576R variant has been shown to interact with other genetic variants to influence atopic disease risk, notably the I75V variant (rs1805010). The combination of Q576R and I75V, specifically the V75/R576 haplotype, leads to an enhanced sensitivity of the IL-4Rα to IL-4 and a stronger association with allergic asthma compared to each individual variant. The association of R576 with the disease is even dependent on the co-existence with V75 variant.26 While no straightforward evidence was documented regarding the epistatic interactions between Q576R and other IL-4R alpha polymorphisms, their known involvement in modulating IL-4R alpha function and their association with atopic diseases suggests the potential for such interactions. Given the complexity of signaling within the Th2 immune pathway, epistatic interactions between the IL-4Rα Q576R polymorphism and genetic variants in related genes are plausible. Evidence shows that gene–gene interactions among IL-4, IL-4Rα, and STAT6 markedly increase atopic dermatitis risk in Egyptian children.21 Besides epistasis, Vietnam’s tropical climate, characterized by high humidity and pollution, may exert selective pressures or interact with Th2-related variants like rs1801275 differently than in Western or even neighboring Asian settings.27 These differences underscore the need for region-specific genetic studies to elucidate AD pathogenesis beyond broad ethnic comparisons.
The G allele (R576) was strongly linked to increased disease severity based on a multi-model genetic analysis (additive, dominant, recessive, and allelic). While rs1801275 did not influence AD onset in the Vietnamese population, its impact on severity was pronounced, as the G allele frequency was similar between patients (17.26%) and controls (20.42%) (p = 0.315). G allele carriers had higher SCORAD scores, particularly in the dominant model (AG+GG vs AA: OR = 4.67, p = 0.005) and additive model (AA vs AG vs GG: OR = 4.52, p = 0.005) compared to A allele ones. In a report of 190 asthmatic inner-city children with AD and 1116 White patients with AD enrolled in the Atopic Dermatitis Research Network, IL-4Rα R576 variant was associated with higher frequency of nighttime itching and Rajka-Langeland score, respectively.10 IL-4 and IL-13 are key Th2 cytokines that signal through the IL-4Rα subunit, which is shared by both the type I IL-4 receptor (IL-4R) and the type II IL-4 receptor (IL-4R/IL-13R). These receptors activate downstream signaling pathways, including the JAK-STAT pathway, leading to the production of IgE and the differentiation of naive T cells into Th2 cells.10,28 The rs1801275 variant in the IL-4Rα gene is associated with increased signaling efficiency in response to IL-4 and IL-13, leading to enhanced Th2 cytokine production and IgE synthesis.10,29 This heightened signaling can exacerbate allergic inflammation and contribute to the severity of atopic diseases. The association of other genetic factors and AD severity has been documented, remarkedly the FLG gene variants. The presence of FLG mutations has been consistently linked to increased disease severity in atopic dermatitis. Studies have shown that up to 40% of European patients with moderate to severe AD carry at least one FLG null mutation.30 Four variants (R1798X, R501X, S126X, and S761fs) of FLG drove the association with SCORAD score, and higher variant load was associated with greater AD severity over time.31 These findings support the role of rs1801275 as a potential predictive marker for AD severity, aiding risk stratification and personalized treatment, particularly in the context of dupilumab therapy inhibiting the IL-4Rα signaling pathway.32
However, limitations exist. The sample size (113 cases, 213 controls) met power requirements but showed only a 3% allele frequency difference, far below the hypothesized 15%, suggesting insufficient power to detect rs1801275’s role in AD onset, though consistent with lower G-allele prevalence in Asians (~20%).14 This did not affect severity assessments within patients. The rarity of GG genotypes (n = 2) weakened recessive model analysis (p = 0.723), reducing statistical power regarding the importance of sample size in genetic studies.33 Factors that could have an impact on disease clinical manifestations represented by SCORAD were under-explored due to the cross-sectional study design and the lack of patient geographical distribution data.11 Lastly, studying only rs1801275 overlooks other IL-4Rα or Th2 pathway variants, critical in AD’s multifactorial nature.7
Future research should prioritize larger, multicenter cohorts to confirm rs1801275’s severity association, particularly for GG genotypes, and adopt longitudinal designs to track progression and management response, notably under IL-4Rα-targeted treatments. Measuring Th2 cytokines (IL-4, IL-13) in G allele carriers could clarify biological mechanisms while exploring gene-environment interactions—such as role of pollution and humidity levels in Vietnam. These steps could solidify rs1801275’s utility as a biomarker, advancing personalized AD management in Southeast Asia.
Conclusion
Our study provides essential preliminary data on the rs1801275 variant in the Vietnamese population. The analysis on four genetic models suggested a potential role of rs1801275 in atopic dermatitis severity rather than disease susceptibility. These findings align with previous studies and lay the foundation for further research on genetics and personalized treatment in Southeast Asia, particularly in the context of a diverse population and limited healthcare resources.
Disclosure
The authors report no conflicts of interest in this work. The authors alone are responsible for the content and writing of the paper.
References
1. Hadi HA, Tarmizi AI, Khalid KA, Gajdacs M, Aslam A, Jamshed S. The Epidemiology and Global Burden of Atopic Dermatitis: a Narrative Review. Life. 2021;11(9):936. doi:10.3390/life11090936
2. Huyen NTH, Hong PTM. To investigate the prevalence of Atopic Dermatitis (AD) and factors related to severity of AD in children from 2 months to 5 years in district I, Ho Chi Minh City. Ho Chi Minh City J Med. 2016;20(1):18–24.
3. Khai HHH, Hanh KTH, Lan PT. Assessment of atopic dermatitis status and some related factors of students of Ha Noi medical university branch in Thanh Hoa. Viet Nam Med J. 2024;538(3):1.
4. Guttman-Yassky E, Renert-Yuval Y, Brunner PM. Atopic dermatitis. Lancet. 2025;405(10478):583–596. doi:10.1016/S0140-6736(24)02519-4
5. Abdel-Mageed HM. Atopic dermatitis: a comprehensive updated review of this intriguing disease with futuristic insights. Inflammopharmacol. 2025;33(3):1161–1187. doi:10.1007/s10787-025-01642-z
6. Dubin C, Del Duca E, Guttman-Yassky E. The IL-4, IL-13 and IL-31 pathways in atopic dermatitis. Expert Rev Clin Immunol. 2021;17(8):835–852. doi:10.1080/1744666X.2021.1940962
7. Paternoster L. Genetic landscape of atopic dermatitis. Curr Opin Allergy Clin Immunol. 2024;24(5):409–415. doi:10.1097/ACI.0000000000001005
8. Tachdjian R, Mathias C, Al Khatib S, et al. Pathogenicity of a disease-associated human IL-4 receptor allele in experimental asthma. J Exp Med. 2009;206(10):2191–2204. doi:10.1084/jem.20091480
9. Massoud AH, Charbonnier L-M, Lopez D, Pellegrini M, Phipatanakul W, Chatila TA. An asthma-associated IL-4R variant exacerbates airway inflammation by promoting conversion of regulatory T cells to TH17-like cells. Nature Med. 2016;22(9):1013–1022. doi:10.1038/nm.4147
10. Yang B, Wilkie H, Das M, et al. The IL-4Ralpha Q576R polymorphism is associated with increased severity of atopic dermatitis and exaggerates allergic skin inflammation in mice. J Allergy Clin Immunol. 2023;151(5):1296–1306e7. doi:10.1016/j.jaci.2023.01.011
11. Cheng J, Wu JJ, Han G. Epidemiology and Characterization of Atopic Dermatitis in East Asian Populations: a Systematic Review. Dermatol Ther. 2021;11(3):707–717. doi:10.1007/s13555-021-00516-w
12. Alenazy AE, Ym A, Si A, Alrohaimi NH. Uses of Dupilumab in Dermatology: a Systematic Review of the Randomized Controlled Trials. Dermato Rev. 2024;5(5):e70005. doi:10.1002/der2.70005
13. Hanifin JM, Rajka G. Diagnostic Features of Atopic Dermatitis. Acta Dermato-Venereol. 1980;60(92):44–47. doi:10.2340/00015555924447
14. Information NCfB. dbSNP: rs1801275. National Library of Medicine (US), National Center for Biotechnology Information. Available from: https://www.ncbi.nlm.nih.gov/snp/rs1801275.
15. Kulthanan K, Tuchinda P, Nitiyarom R, et al. Clinical practice guidelines for the diagnosis and management of atopic dermatitis. Asian Pac J Allergy Immunol. 2021;39(3):145–155. doi:10.12932/AP-010221-1050
16. Severity Scoring Of Atopic Dermatitis: The SCORAD Index. Consensus Report of the European Task Force on Atopic Dermatitis. Dermatology. 1993;186(1):23–31. doi:10.1159/000247298
17. Thoi LT, Dat NQ, Ha NM, Tuan NHN. Establishing a Real-time PCR SYBR procedure for detecting rs1801275 variant in the IL4-Rα gene associated with the atopic dermatitis. Viet Nam Med J. 2024;534(1B):2.
18. Chong AC, Visitsunthorn K, Ong PY. Genetic/Environmental Contributions and Immune Dysregulation in Children with Atopic Dermatitis. J Asthma Allergy. 2022;15:1681–1700. doi:10.2147/JAA.S293900
19. Georas SN, Guo J, De Fanis U, Casolaro V. T-helper cell type-2 regulation in allergic disease. Eur Respir J. 2005;26(6):1119–1137. doi:10.1183/09031936.05.00006005
20. Scibiorek M, Mthembu N, Mangali S, et al. IL-4Rα signalling in B cells and T cells play differential roles in acute and chronic atopic dermatitis. Sci Rep. 2023;13(1):144. doi:10.1038/s41598-022-26637-6
21. Hussein YM, Shalaby SM, Nassar A, Alzahrani SS, Alharbi AS, Nouh M. Association between genes encoding components of the IL-4/IL-4 receptor pathway and dermatitis in children. Gene. 2014;545(2):276–281. doi:10.1016/j.gene.2014.04.024
22. Oiso N, Fukai K, Ishii M. Interleukin 4 receptor alpha chain polymorphism Gln551Arg is associated with adult atopic dermatitis in Japan. Br J Dermatol. 2000;142(5):1003–1006. doi:10.1046/j.1365-2133.2000.03485.x
23. Isidoro-García M, Davila I, Moreno E, Laffond E, Lorente F, González-Sarmiento R. IL-4RA gene polymorphism (Q576R) is associated with higher total IgE levels in Spanish patients with family history of atopy. Med Clin. 2005;124(6):211–212. doi:10.1157/13071760
24. Tanaka K, Sugiura H, Uehara M, Hashimoto Y, Donnelly C, Montgomery DS. Lack of association between atopic eczema and the genetic variants of interleukin-4 and the interleukin-4 receptor alpha chain gene: heterogeneity of genetic backgrounds on immunoglobulin E production in atopic eczema patients. Clin Exp Allergy. 2001;31(10):1522–1527. doi:10.1046/j.1365-2222.2001.01205.x
25. Kim J, Ahn K. Atopic dermatitis endotypes: knowledge for personalized medicine. Current Opinion in Allergy & Clinical Immunology. 2022;22(3):153–159. doi:10.1097/ACI.0000000000000820
26. Risma KA, Wang N, Andrews RP, et al. V75R576 IL-4 receptor alpha is associated with allergic asthma and enhanced IL-4 receptor function. J Immunol. 2002;169(3):1604–1610. doi:10.4049/jimmunol.169.3.1604
27. Shigeru S. Pollutants and climatic conditions related to the childhood asthma and atopic dermatitis. World J Biol Pharm Health Sci. 2022;9(1):027–038. doi:10.30574/wjbphs.2022.9.1.0025
28. Al-Muhsen S, Vazquez-Tello A, Alzaabi A, Al-Hajjaj MS, Al-Jahdali HH, Halwani R. IL-4 receptor alpha single-nucleotide polymorphisms rs1805010 and rs1801275 are associated with increased risk of asthma in a Saudi Arabian population. Ann Thorac Med. 2014;9(2):81–86. doi:10.4103/1817-1737.128849
29. Brian W, Brenna L, Anne C, et al. The IL-4Rα Q576R Polymorphism Promotes Th2 Skewing In Atopic Dermatitis Patients. J Allergy Clin Immunol. 2024;153(2):AB255. doi:10.1016/j.jaci.2023.11.816
30. Brown SJ, Elias MS, Bradley M. Genetics in Atopic Dermatitis: historical Perspective and Future Prospects. Acta Derm Venereol. 2020;100(12):adv00163. doi:10.2340/00015555-3513
31. Huffaker MF, Kanchan K, Bahnson HT, et al. Epidermal differentiation complex genetic variation in atopic dermatitis and peanut allergy. J Allergy Clin Immunol. 2023;151(4):1137–1142e4. doi:10.1016/j.jaci.2022.11.008
32. Harb H, Chatila TA. Mechanisms of Dupilumab. Clin Exp Immunol. 2020;50(1):5–14.
33. Park JH, Gail MH, Weinberg CR, et al. Distribution of allele frequencies and effect sizes and their interrelationships for common genetic susceptibility variants. Proc Natl Acad Sci U S A. 2011;108(44):18026–18031. doi:10.1073/pnas.1114759108
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




