Back to Journals » Journal of Inflammation Research » Volume 18
Heightened Innate Immune and Inflammatory Gene Expression in the Skin of Severe COVID-19 Cases
Authors Fernandes IG, Ferreira FM, De Sousa ESA , Pietrobon AJ, Teixeira FME, Ramos YÁL, Yendo TM, Pereira NZ, Sotto MN , Duarte-Neto AN , Orfali RL, Aoki V, Da Silva LFF, Duarte AJDS, Sato MN
Received 10 January 2025
Accepted for publication 28 June 2025
Published 12 July 2025 Volume 2025:18 Pages 9119—9128
DOI https://doi.org/10.2147/JIR.S516737
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Anish R. Maskey
Iara Grigoletto Fernandes,1 Frederico Moraes Ferreira,2 Emanuella Sarmento Alho De Sousa,1,3 Anna Julia Pietrobon,1 Franciane Mouradian Emidio Teixeira,1 Yasmim Álefe Leuzzi Ramos,1 Tatiana Mina Yendo,1 Natalli Zanete Pereira,1 Mirian Nacagami Sotto,4 Amaro Nunes Duarte-Neto,4 Raquel Leão Orfali,1 Valeria Aoki,1 Luiz Fernando Ferraz Da Silva,4 Alberto José da Silva Duarte,1 Maria Notomi Sato1
1Laboratory of Medical Investigation 56, Department of Dermatology, University of São Paulo Medical School, São Paulo, Brazil; 2Cellular, Genetic, and Molecular Nephrology Laboratory (LIM-29), University of São Paulo Medical School, São Paulo, Brazil; 3Department of Immunology, Institute of Biomedical Sciences, University of São Paulo, São Paulo, Brazil; 4Laboratory of Medical Investigation 50, Department of Pathology, University of São Paulo Medical School, São Paulo, Brazil
Correspondence: Maria Notomi Sato, Laboratory of Medical Investigation in Dermatology and Immunodeficiencies (LIM-56), Department of Dermatology, Faculdade de Medicina, Universidade de Sao Paulo, Sao Paulo, SP, 05403-000, Brazil, Tel +55 11 30617499, Email [email protected]
Purpose: Cutaneous manifestations of SARS-CoV-2 infection exhibit significant variability, yet the role of innate immune responses in the skin of COVID-19 patients remains poorly understood. In this study, we investigated the transcriptomic profile of skin samples from patients who succumbed to COVID-19.
Patients and Methods: Skin autopsies from COVID-19 patients with post-mortem time of less than 20 hours were obtained from University of São Paulo Medical School Hospital and healthy skin samples, were submitted to RNA sequencing analysis. Validation of differentially expressed genes (DEGs) was performed by real-Time PCR.
Results: Our analysis revealed markedly elevated expression of type I interferon (IFN)-inducible antiviral factors, antioxidant enzymes, and components of several cytokine-signaling pathways in COVID-19 skin samples compared to healthy controls. SARS-CoV-2 infection robustly induced numerous interferon-stimulated genes (ISGs), IFITs, IRF, S100 family with a notable enrichment of those associated with antiviral and inflammatory responses. Moreover, the presence of counter-regulatory factors such as SOCS3 and NFKBIA indicate the involvement of anti-inflammatory mechanisms in the skin. Furthermore, deconvolution data indicated increased presence of macrophages other nucleated cells in vessels in the skin of COVID-19 patients, highlighting the involvement of innate immune mechanisms.
Conclusion: The results revealed cutaneous alterations in the expression of genes associated with innate immunity and inflammation factors. This suggests that, unlike tissues with viral tropism, the skin is enriched with antiviral factors to defend against SARS-CoV-2. This information could be useful for developing specific antiviral therapies.
Keywords: COVID-19, SARS-CoV-2, skin, RNA-Seq, innate immunity
Introduction
The cutaneous manifestations associated with COVID-19 vary widely and can be broadly classified into two main categories: inflammatory reactions (eg, morbilliform (measles-like) rash, vascular rashes and vesicular rash) and vascular-origin lesions (eg, chilblain-like rashes, petechiae/purpura, and livedo racemosa-like patterns).1,2 Vesicular eruptions appear early in the course of the disease (15% before other symptoms). The pseudo‐chilblain pattern frequently appears late in the evolution of the COVID‐19 disease (59% after other symptoms), while the rest tend to appear with other symptoms of COVID‐19.3 Despite numerous reports of diverse cutaneous manifestations linked to SARS-CoV-2 infection, dermatologists treating COVID-19 patients at the University of São Paulo Medical School Hospital—a major reference center and one of the largest university hospitals in Latin America confirmed that was not possible to attribute with certainty that any cutaneous lesions had a direct correlation to the SARS-CoV-2 infection in 86 patients confirmed with COVID-19.4 Instead, the dermatological presentations observed mirrored those commonly encountered prior to the pandemic, with fungal infections being the most frequent, followed by drug eruptions and viral infections. This underscores the need for further molecular investigations to unravel the immunological changes occurring in the skin during COVID-19. In same line, it has been verified a lack of association between chilblains and SARS-CoV-2 infection.5
SARS-CoV-2 enters host cells via the angiotensin-converting enzyme 2 (ACE2) receptor and TMPRSS2 protease, which are expressed in multiple tissues, including the lungs, kidneys, colon, heart, testicles, and skin.6 Despite this, studies have found no virally induced cytopathic alterations or intranuclear inclusions in the skin,7 nor detectable SARS-CoV-2 viral load in skin biopsy specimens.8,9 Nevertheless, upon viral entry, after the latent period, depending on the variant, such as Delta, the latency period was 3.2 days.10 After this, a cascade of activation of innate immunity is rapidly activated, including the recruitment of dendritic cells, macrophages, and monocytes. These immune cells adhere to infected cells and release type I interferons (IFNs), which act as danger signals, along with pro-inflammatory and antiviral cytokines.
Understanding the molecular immunological events in the skin during SARS-CoV-2 infection requires a focus on innate immunity, which involves pattern recognition receptors (PRRs), Toll-like receptor (TLR) signaling, and cytokine-mediated antiviral responses. To determine the extent to which the skin is activated in response to SARS-CoV-2. If activation does occur, it is crucial to identify the controlling factors.
In this study, we conducted a transcriptomic analysis of skin samples from patients with acute COVID-19 as depicted in Figure 1A. Our findings reveal an integrated gene expression profile encompassing antiviral, antioxidant, and innate immune responses in the skin, shedding light on the complex host-pathogen interactions in this compartment.
Materials and Methods
Patients’ Enrollment
Skin samples used in this study were obtained from patients who succumbed to severe SARS-CoV-2 infection while hospitalized in the Intensive Care Unit of Hospital das Clínicas, São Paulo. Post-mortem biopsies were collected in the thigh region using a 5 mm punch, by the Death Verification Service. All patients were diagnosed with COVID-19 through nasopharyngeal detection of SARS-CoV-2 RNA using reverse transcriptase polymerase chain reaction (RT-PCR).
For RNA sequencing, we included skin samples from eight COVID-19 patients (4 men and 4 women), aged 32–70 years, who died between April and June 2020 (Supplementary Table 1). Additional skin samples from COVID-19 patients (n=10, 3 men and 7 women, aged 26–76 years) were used for real-time PCR analysis. At the time, vaccines against SARS-CoV-2 were unavailable. Post-mortem skin biopsies were collected within 20 hours of death from the same anatomical region for all patients. Post-mortem skin biopsies were taken up to 20 hours after death due to complications arising from a diagnosis of SARS-CoV-2. The exclusion criteria were cancer, HIV-1 infection, and others skin diseases.
As controls, skin biopsies were obtained from healthy donors (n=5) prior to the COVID-19 pandemic. These control samples were collected from Caucasian individuals of both sexes, aged 35–70 years. The autopsy was carried out with the authorization of the relatives, who signed an informed consent form approved by the National Research Ethics Committee (CONEP). The study was approved by the ethics committee of the HC-FMUSP (CAAE 30364720.0.0000.0068) and conducted following the Declaration of Helsinki.
Transcriptomic Analysis
Total RNA was extracted from autopsy-derived skin specimens using the RNeasy Plus Mini Kit (Qiagen, Hilden, Germany), following the manufacturer’s instructions. Tissue homogenization was performed with a TissueRuptor (Qiagen). RNA integrity was assessed using the TapeStation system (Agilent, Santa Clara, CA, USA).
Transcriptomic profiling was carried out using the Illumina TruSeq Stranded mRNA Library Prep Kit (Illumina, San Diego, CA, USA) at the FMUSP Large-Scale Sequencing Laboratory (SELA). Sequencing adapters were removed, and quality control of raw reads was performed using FastQC.11 Reads were aligned to the reference human genome (GRCh38.p13, version 107) using the SUBREAD package12 and quantified with FEATURECOUNTS.13
Differentially expressed genes (DEGs) were identified using the DESeq2 package from R/Bioconductor,14 with statistical significance determined at an adjusted p-value ≤ 0.05 (Benjamini-Hochberg correction). Functional enrichment analyses were conducted with the FGSEA package15 using KEGG16 and GO Biological Process17 databases. Protein-protein interaction networks were constructed using Cytoscape18 with data from the InnateDB database.19
Gene Expression by Real-Time PCR
Total RNA was quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific). Complementary DNA (cDNA) was synthesized using the iScript™ Reverse Transcription Kit (Bio-Rad, Hercules, CA, USA). Real-time PCR was performed with SYBR Green (Applied Biosystems, Waltham, MA, USA) and primers specific to the target genes (primer sequences listed in Supplementary Table 2). Amplification was carried out on a 7500 Real-Time PCR System (Applied Biosystems), and data analysis was conducted using 7500 software v2.0.6 (Applied Biosystems) based on the ΔΔCt method.20
Statistical Analysis
Statistical comparisons between groups were performed using the Mann–Whitney test, with significance set at p ≤ 0.05. Detection of outlier samples, normalization, statistical tests and identification of DEGs were carried out using the package R DESEQ2 from the Bioconductor repository. The p-values were corrected at using the Benjamini-Hochberg method. Genes were considered differentially expressed for p-adj values ≤ 0.05 and absolute fold-change values, FC ≥ 1.5.
Results
Characteristics of the COVID-19 Patients and Healthy Controls
A group of 8 patients who died due to COVID-19 complications (CV) from April to June 2020, composed of 4 men and 4 women, aged between 32 to 76 years (mean = 55,25), was evaluated (Supplementary Table 1). All cases configure severity and death, and comorbidities were identified in almost all individuals. As a control group, we used skin samples from 5 healthy donors (HD), composed of 2 men and 3 women, aged between 29 and 70 years (mean = 62). HD skin samples were collected in 2019, before the COVID-19 pandemic. No comorbidities or infectious diseases were present in this group.
Transcriptomic Profile of Skin Samples
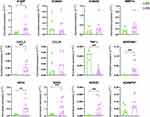
Skin samples were processed for RNA extraction, followed by library construction and sequencing. Computational analyses were performed to identify differentially expressed genes (DEGs), enriched molecular pathways, and gene interaction networks. Gene expression of the most important DEGs were validated using real-time PCR (Figure 1A).
Unsupervised z-score heatmap of normalized counts revealed a distinct transcriptomic profile between COVID-19 (CV) and healthy control (CT) skin samples (Figure 1B). This showed 150 upregulated and 150 downregulated genes. The more important genes related to innate immunity were labelled. A total list of genes can be found in the Supplementary Table 3. A volcano plot demonstrated with total genes, that 1188 upregulated and 803 downregulated genes in the CV group compared to the CT group (Figure 2). Principal component analysis (PCA) showed that the great majority of the samples grouped closely within their respective cohorts, except for one perhaps mild CV sample (Figure 2).
Enrichment molecular pathway analysis revealed upregulation of 25 pathways associated with inflammatory responses in the CV group compared to control group. These pathways included “regulation of inflammatory response”, “response to molecules of bacterial origin”, “leukocyte chemotaxis”, “signaling by interleukins”, “response to oxidative stress”, “matrix organization”, and the “JAK-STAT signaling pathway”, all of which are linked to innate immunity and inflammation (Figure 3A). Moreover, pathways analyzed included response to oxidative stress as “reactive oxygen species metabolic response” and “response to oxidative stress”. This finding was in line with downregulation of pathways related to controlling of inflammatory process, such “positive regulation of signaling”, “regulation of cell differentiation”, “negative regulation of response to stimulus”, “homeostatic process” (Figure 3B).
Over-representation analysis revealed downregulated molecular pathways in CV samples included those related to the skin matrix, such as “matrisome”, “collagen-containing extracellular matrix”, “epidermis development”, “regulation of cell differentiation”, and “connective tissue development” (Figure 3B).
The gene interaction network revealed several upregulated DEGs (pink nodes) associated with antiviral responses, including interferon-stimulated genes (ISGs) such as IFIT1, IFIT2, IFIT3, and interferon-regulatory factors IRF3 and IRF7. Additional antiviral genes included members of the S100 protein family, such as S100P, S100A7, S100A8, and S100A9. Cytokine and cytokine-related factors were also prominent, including STAT2, STAT3, SOCS3, NF-κB2, RELB, NFKBIA, MyD88, IL-6, CCL20, and CXCL2 (Figure 4). Some factors were downregulated (blue nodes), such as TNF, a multifunctional proinflammatory cytokine, and MAVS (mitochondrial antiviral signaling protein), which regulates IFN-β expression and contributes to antiviral innate immunity (Figure 4). Figure 5 shows the selected DEGs for validation in skin samples, which showed upregulation of S100P, GPX3, SOD2, SOCS3, SERPINE1, and CXCL2, alongside downregulation of TNF-α in the skin of COVID-19 patients compared to the CT group.
In addition, deconvolution analysis of immune cell types revealed an increased presence of macrophages and reduced dendritic cells in the skin tissue of COVID-19 patients, likely reflecting their migratory status (Supplementary Figure 1). Eosinophils were only present inside the dermal perivascular vessel, not in the tissue infiltration. There is mild lymphohistiocytic infiltration in skin tissue (see Supplementary Figure 2).
Discussion
The gene expression profile of the skin in COVID-19 patients revealed significant upregulation of innate immune factors including antiviral components, antioxidant defenses, and inflammatory markers. Although the skin is not a primary target organ for SARS-CoV-2, it engages protective mechanisms that may prevent severe cutaneous lesions. Network analysis highlighted the involvement of innate antiviral immune factors such as Mx1, APOBEC3G, IRF3, IRF7, IRF9, DDX58, IFIT3, and IFIT1. These genes are associated with antiviral responses triggered by interferon signaling, indicating a robust cutaneous antiviral response. Additionally, interferon-induced transmembrane (IFITM) proteins, known to be upregulated in multiple tissues such as the lung, gut, heart, and brain,21 were also expressed in the skin, further underscoring its protective role. Despite the absence of detectable viral load in most cases (present in only two samples), all skin samples exhibited upregulation of antiviral genes.
In terms of innate immunity, the differentially expressed genes found in COVID-19 skin samples included members of the S100 protein family, such as S100A8, S100A9, and S100A7. These proteins, particularly S100A8/A9, are known alarmins that mediate host pro-inflammatory responses and have been identified as biomarkers of COVID-19 severity.22 Consistent with findings in nasal swabs, the upregulation of S100 genes was associated with disease severity.23 In the skin, S100A8/A9 acts as a ligand for TLR4 and RAGE, promoting NF-κB signaling and inducing pro-inflammatory cytokines such as IL-6, which was notably upregulated in COVID-19 skin samples. This contrasts with the downregulation of TNF-α, a cytokine often implicated in the cytokine storm of severe COVID-19 cases.24 The reduced expression of TNF-α in the skin may represent a counter-regulatory mechanism to limit excessive inflammation, possibly mediated by the upregulation of SOCS3, a known negative feedback inhibitor of cytokine signaling, and also includes NFKBIA which encodes a protein known as IκBα (NF-κB alpha inhibitor). These findings align with prior evidence of cross-regulation between TNF-α and type I IFNs triggering,25 where TNF-α has been shown to suppress type I IFN production and pDC activity, while IFN-β can inhibit TNF-α secretion. Given that TNF-α may suppress an important antiviral factor such as type I IFN, the cutaneous reduction in TNF-α is timely.
Over-representation analysis obtained with the downregulated genes related with impaired development of the extracellular epidermis and connective tissue and influence the dynamic repair of skin tissue in skin of severe cases of COVID-19. However, the upregulation of CXCL2 plays a role in the stages of haemostasis, inflammation, proliferation and remodeling in the skin. This finding suggests that, rather than acting as an eosinophil/neutrophil chemoattractant, CXCL2 acts more in skin remodeling. This is particularly relevant given that skin infiltration in samples from patients with SARS-CoV-2 was primarily composed of lymphocytes and histiocytes (macrophages).
The number of downregulated genes related to innate immunity in the skin was much smaller than the number of upregulated genes. Such as, TNF-α, mitochondrial antiviral signaling protein (MAVS) and WNT2. MAVS plays a key role in the antiviral response. MAVS is activated in response to viral infection by RIG-I-like receptors that recognize viral RNA, mediating the activation of NF-κB and interferon-regulatory factors, as well as the induction of interferons, in response to viral infection. It is required for interferon induction in dendritic cells (DCs).26 This observation aligns with previous reports that SARS-CoV-2 nucleocapsid protein inhibits type I IFN responses by interacting with MAVS, thereby suppressing host antiviral defenses. However, it remains unclear whether the downregulation of MAVS in the skin reflects a specific viral evasion strategy. The Wnt/β-catenin signaling pathway inhibits the replication of SARS-CoV-2 and other pathogenic RNA viruses in vitro.27 Furthermore, the Wnt/β-catenin signaling pathway strongly suppresses peroxisome biogenesis. These are intracellular organelles containing enzymes for beta-oxidation, and they are depleted from cells and tissues during SARS-CoV-2 infection. This depletion may be expected to impair the IFN response.28
Despite the downregulation of MAVS, our findings showed that some first-line defense antioxidants, such as superoxide dismutase (SOD) and glutathione peroxidase (GPX), are upregulated in the skin. These genes play critical roles in mitigating oxidative stress, with SOD2 converting reactive oxygen species (ROS) to hydrogen peroxide and GPX3 metabolizing hydrogen peroxide and lipid hydroperoxides toxins.29 This robust antioxidant response highlights the importance of oxidative stress mitigation in SARS-CoV-2 pathogenesis and suggests potential therapeutic targets.
Another notable upregulated DEG was SERPINE1, which encodes plasminogen activator inhibitor 1 (PAI-1). Upregulation of PAI-1 may counteract the thrombosis frequently observed in COVID-19 by modulating fibrinolysis. SERPINE 1 is required to stimulate keratinocyte migration during the repair of cutaneous injuries.30 A polymorphism in the human SERPINE1 gene increases influenza A virus susceptibility in vitro.31 Furthermore, the ability of SERPINE1 to inhibit TMPRSS2 has been demonstrated, resulting in a reduction in SARS-CoV-2 entry and infection.32 We observed that protease inhibitors such as SERPINE 1, was observed in heatmap as DEGs, in the expressions in skin from COVID-19, have shown to be valuable component of the antiviral innate immunity.
Analysis of immune cell composition revealed an increased presence of macrophages in the skin of COVID-19 patients, along with a reduction in DCs. The decreased number of DCs in the skin may reflect their migration to the draining lymph nodes, where antigen-specific adaptive immunity occurs. Conversely, eosinophils were only present in the perivascular dermal vessels and not in the tissue infiltrate, which was found to be mild lymphohistiocytic infiltration of the skin.
Conclusion
SARS-CoV-2 triggers a significant innate immune response in the skin, characterised by the upregulation of antiviral, inflammatory and antioxidant pathways. Several genes related to innate immunity are upregulated, including members of the S100 family, IFITs, ISGs, pro-inflammatory cytokines, and chemokines, indicating a robust cutaneous antiviral response but also with inflammatory potential. In this line, a counter-regulatory response is induced, by genes such as SOCS3, which controls cytokine levels, and NFKBIA, which controls NF-κB activity. These genes may, partly, control the inflammatory status in the skin. The severe COVID-19 patient showed intense innate immunity enriched with antiviral factors. This could explain why they did not exhibit significant cutaneous lesions. Further investigation into the interaction of immune pathways in the skin could lead to the development of new therapeutic strategies for controlling the inflammation associated with the disease.
Ethical Approval
The study was approved by the committee for Ethics Committee of the Hospital das Clínicas da Faculdade de Medicina da Universidade de São Paulo–HC-FMUSP (CAAE 30800520.7.0000.0068-2020).
Acknowledgments
The authors are extremely grateful for the participation of skin samples from volunteers, and the autopsy of skin samples.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Laboratório de Investigação Médica, Unidade 56, Department of Dermatology, School of Medicine, University of São Paulo, Brazil; Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) under Grant [No. 2019/25119-7], Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) under Grant [No. 88887.503842/2020-00].
Disclosure
The author(s) report no conflicts of interest in this work.
References
1. Martora F, Villani A, Battista T, Fabbrocini G, Potestio L. COVID‐19 vaccination and inflammatory skin diseases. J Cosmet Dermatol. 2022. doi:10.1111/jocd.15414
2. Zaborska M, Chruszcz M, Sadowski J, et al. The most common skin symptoms in young adults and adults related to SARS-CoV-2 virus infection. Arch DermatolRes. 2024;316(6):292. doi:10.1007/s00403-024-02991-5
3. Galván Casas C, Català A, Carretero Hernández G, et al. Classification of the cutaneous manifestations of COVID-19: a rapid prospective nationwide consensus study in Spain with 375 cases. Br J Dermatol. 2020;183(1):71–77. doi:10.1111/bjd.19163
4. Avancini J, Miyamoto D, Arnone M, et al. Absence of specific cutaneous manifestations of severe acute respiratory syndrome coronavirus 2 in a reference center in Brazil. J Am Acad Dermatol. 2021;84(1):e67. doi:10.1016/j.jaad.2020.09.030
5. Gehlhausen JR, Little AJ, Ko CJ, et al. Lack of association between pandemic chilblains and SARS-CoV-2 infection. Proc Natl Acad Sci U S A. 2022;119(9). doi:10.1073/pnas.2122090119
6. Dong M, Zhang J, Ma X, et al. ACE2, TMPRSS2 distribution and extrapulmonary organ injury in patients with COVID-19. Biomed Pharmacother. 2020;131:110678. doi:10.1016/j.biopha.2020.110678
7. Amatore F, Macagno N, Mailhe M, et al. SARS‐CoV‐2 infection presenting as a febrile rash. J Eur Acad Dermatol Venereol. 2020;34(7):e304. doi:10.1111/jdv.16528
8. Ahouach B, Harent S, Ullmer A, et al. Cutaneous lesions in a patient with COVID‐19: are they related? Br J Dermatol. 2020;183(2):e31–e31. doi:10.1111/bjd.19168
9. Fernandez‐Nieto D, Ortega‐Quijano D, Jimenez‐Cauhe J, et al. Clinical and histological characterization of vesicular COVID‐19 rashes: a prospective study in a tertiary care hospital. Clin Exp Dermatol. 2020;45(7):872–875. doi:10.1111/ced.14277
10. Park SW, Sun K, Abbott S, et al. Inferring the differences in incubation-period and generation-interval distributions of the Delta and Omicron variants of SARS-CoV-2. Proc Natl Acad Sci U S A. 2023;120(22):e2221887120. doi:10.1073/pnas.2221887120
11. Wingett SW, Andrews S. FastQ screen: a tool for multi-genome mapping and quality control. F1000Research. 2018;7:1338. doi:10.12688/f1000research.15931.1
12. Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41(10):e108–e108. doi:10.1093/nar/gkt214
13. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi:10.1093/bioinformatics/btt656
14. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi:10.1186/s13059-014-0550-8
15. Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, Sergushichev A. Fast gene set enrichment analysis. biorxiv. 2016;2016:060012.
16. Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023;51(D1):D587–D592. doi:10.1093/nar/gkac963
17. Consortium GO. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32(suppl_1):D258–D261. doi:10.1093/nar/gkh036
18. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
19. Breuer K, Foroushani AK, Laird MR, et al. InnateDB: systems biology of innate immunity and beyond—recent updates and continuing curation. Nucleic Acids Res. 2013;41(D1):D1228–D1233. doi:10.1093/nar/gks1147
20. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
21. Hachim MY, Al Heialy S, Hachim IY, et al. Interferon-induced transmembrane protein (IFITM3) is upregulated explicitly in SARS-CoV-2 infected lung epithelial cells. Front Immunol. 2020;11:1372. doi:10.3389/fimmu.2020.01372
22. Mellett L, Khader SA. S100A8/A9 in COVID-19 pathogenesis: impact on clinical outcomes. Cytokine Growth Factor Rev. 2022;63:90–97. doi:10.1016/j.cytogfr.2021.10.004
23. Biji A, Khatun O, Swaraj S, et al. Identification of COVID-19 prognostic markers and therapeutic targets through meta-analysis and validation of Omics data from nasopharyngeal samples. EBioMedicine. 2021;70:1.
24. Del Valle DM, Kim-Schulze S, Huang -H-H, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 2020;26(10):1636–1643. doi:10.1038/s41591-020-1051-9
25. Palucka AK, Blanck J-P, Bennett L, Pascual V, Banchereau J. Cross-regulation of TNF and IFN-α in autoimmune diseases. Proc Natl Acad Sci. 2005;102(9):3372–3377. doi:10.1073/pnas.0408506102
26. Ren Z, Ding T, Zuo Z, Xu Z, Deng J, Wei Z. Regulation of MAVS expression and signaling function in the antiviral innate immune response. Front Immunol. 2020;11:1030. doi:10.3389/fimmu.2020.01030
27. Xu Z, Elaish M, Wong CP, et al. The Wnt/β-catenin pathway is important for replication of SARS-CoV-2 and other pathogenic RNA viruses. Npj Viruses. 2024;2(1):6. doi:10.1038/s44298-024-00018-4
28. Roczkowsky A, Limonta D, Fernandes JP, et al. COVID-19 induces neuroinflammation and suppresses peroxisomes in the brain. Ann Neurol. 2023;94(3):531–546. doi:10.1002/ana.26679
29. Wu JH, Batist G. Glutathione and glutathione analogues; therapeutic potentials. Biochim Biophys Acta. 2013;1830(5):3350–3353. doi:10.1016/j.bbagen.2012.11.016
30. Providence KM, Higgins SP, Mullen A, et al. SERPINE1 (PAI-1) is deposited into keratinocyte migration “trails” and required for optimal monolayer wound repair. Arch Dermatol Res. 2008;300(6):303–310. doi:10.1007/s00403-008-0845-2
31. Dittmann M, Hoffmann HH, Scull MA, et al. A serpin shapes the extracellular environment to prevent influenza A virus maturation. Cell. 2015;160(4):631–643. doi:10.1016/j.cell.2015.01.040
32. Rosendal E, Mihai IS, Becker M, et al. Serine protease inhibitors restrict host susceptibility to SARS-CoV-2 infections. mBio. 2022;13(3):e0089222. doi:10.1128/mbio.00892-22
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
Recommended articles
COVID-19 and Saudi Arabia: Awareness, Attitude, and Practice
Fawzy MS, AlSadrah SA
Journal of Multidisciplinary Healthcare 2022, 15:1595-1618
Published Date: 26 July 2022
Comparison of the Diagnostic Performance of a Rapid Antigen Test with Real-Time Polymerase Chain Reaction for Detection of SARS-CoV-2 Among Patients Diagnosed with COVID-19 at Selected Hospitals in Addis Ababa, Ethiopia
Desalegn Z, Sebre S, Yohannes M, Seman A, Shiferaw W, Ademe M, Biazin H, Firdawoke E, Asemamaw Y, Teka B, Teshome S, Amogne W, Addissie A, Gebrehiwot Y, Kantelhardt E, Abebe T
Infection and Drug Resistance 2022, 15:4299-4305
Published Date: 6 August 2022
A Pilot Study of 0.4% Povidone-Iodine Nasal Spray to Eradicate SARS-CoV-2 in the Nasopharynx
Sirijatuphat R, Leelarasamee A, Puangpet T, Thitithanyanont A
Infection and Drug Resistance 2022, 15:7529-7536
Published Date: 21 December 2022
Distinct Features of Vascular Diseases in COVID-19
Ceasovschih A, Sorodoc V, Shor A, Haliga RE, Roth L, Lionte C, Onofrei Aursulesei V, Sirbu O, Culis N, Shapieva A, Tahir Khokhar MA, Statescu C, Sascau RA, Coman AE, Stoica A, Grigorescu ED, Banach M, Thomopoulos C, Sorodoc L
Journal of Inflammation Research 2023, 16:2783-2800
Published Date: 6 July 2023
Re-Emerging COVID-19: Controversy of Its Zoonotic Origin, Risks of Severity of Reinfection and Management
Chala B, Tilaye T, Waktole G
International Journal of General Medicine 2023, 16:4307-4319
Published Date: 20 September 2023