Back to Journals » Breast Cancer: Targets and Therapy » Volume 17
Heterogeneity Assessment of Breast Cancer Tumor Microenvironment: Multiparametric Quantitative Analysis with DCE-MRI and Discovery of Radiomics Biomarkers
Authors Ma W, Yang L, Zhang Y, Gao Y, Jie H, Huang C
Received 27 March 2025
Accepted for publication 2 July 2025
Published 8 July 2025 Volume 2025:17 Pages 573—581
DOI https://doi.org/10.2147/BCTT.S530834
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Robert Clarke
Wenhui Ma,1 Lu Yang,1 Yu Zhang,1 Yuan Gao,1 Huan Jie,2 Cong Huang1
1Department of Radiology, No. 926 Hospital, Joint Logistics Support Force of PLA, Kaiyuan, Yunnan, 661699, People’s Republic of China; 2Department of Oncology, No. 926 Hospital, Joint Logistics Support Force of PLA, Kaiyuan, Yunnan, 661699, People’s Republic of China
Correspondence: Cong Huang, Department of Radiology, No. 926 Hospital, Joint Logistics Support Force of PLA, Kaiyuan, Yunnan, 661699, People’s Republic of China, Email [email protected]
Abstract: The heterogeneity of the tumor microenvironment (TME) in breast cancer significantly influences therapeutic response and prognosis, yet noninvasive evaluation remains a clinical challenge. Dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI), through multiparametric quantitative analysis (eg, Ktrans, Ve, Kep), enables dynamic characterization of tumor vascularization and perfusion heterogeneity. Concurrently, radiomics technology, leveraging high-throughput feature extraction and machine learning modeling, identifies potential biomarkers associated with TME biological properties. This review systematically examines the integration strategies of DCE-MRI multiparametric quantification and radiomics: first, elucidating the capability of DCE-MRI pharmacokinetic models to quantify microvascular heterogeneity, and delineating radiomics feature screening and predictive model construction based on 3D segmentation. Furthermore, it explores the combined application of these techniques in evaluating angiogenesis, resolving immune microenvironment dynamics, and mapping metabolic heterogeneity, with emphasis on clinical translational evidence in molecular subtype discrimination, treatment response prediction, and prognostic assessment. Key limitations persist in technical standardization (eg, 37% variability in Ktrans values across 1.5T/3.0T systems) and biological interpretability, with fewer than 40% of radiomics features linked to known molecular pathways. Future advancements demand multicenter data harmonization, radiogenomics integration, and digital twin technology to optimize personalized therapeutic navigation systems. This work provides methodological insights and technical innovation pathways for noninvasive TME heterogeneity assessment in breast cancer.
Keywords: breast cancer, tumor microenvironment, heterogeneity, dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI), radiomics, biomarkers, radiogenomics
Introduction
Breast cancer remains the most prevalent malignancy in women worldwide, exhibiting marked heterogeneity in therapeutic response and clinical outcomes that poses critical challenges for treatment optimization.1 This biological diversity originates from the tumor microenvironment (TME) – a dynamically organized ecosystem comprising immune infiltrates, cancer-associated fibroblasts, neo-vasculature, and remodeled extracellular matrix. The spatial-temporal evolution of TME components drives key oncogenic processes including immune evasion, metastatic progression, and therapeutic resistance through complex cellular crosstalk.2 While histopathological analysis remains the diagnostic gold standard, conventional biopsy techniques suffer from inherent limitations: invasive sampling risks complications (2–15% hematoma incidence), spatial sampling bias (78% concordance between biopsy and surgical specimens for receptor status), and inability to capture longitudinal heterogeneity patterns across entire tumor volumes.3
Advanced quantitative MRI techniques have revolutionized noninvasive TME characterization, particularly dynamic contrast-enhanced MRI (DCE-MRI). By modeling contrast agent pharmacokinetics, DCE-MRI enables precise quantification of microvascular permeability (Ktrans) and fractional extracellular-extravascular volume (Ve), parameters that correlate strongly with histopathological microvessel density (r = 0.62–0.79) and collagen deposition (r = 0.54).4 Parallel developments in radiomics provide complementary insights through high-dimensional feature extraction (first-order statistics, texture patterns, wavelet decompositions) from multiparametric MRI data, with emerging capabilities to decode TME biology via machine learning-driven pattern recognition.5 Recent multicenter studies demonstrate the clinical potential of integrated models combining DCE-MRI parameters with radiomic signatures, achieving 82% accuracy in triple-negative subtype prediction (vs 68% for DCE-MRI alone) and 0.89 AUC for neoadjuvant therapy response assessment.6–8 Nevertheless, critical barriers impede clinical translation, including poor inter-scanner reproducibility of quantitative parameters (23–41% coefficient of variation for Ktrans across vendors) and limited biological interpretability of radiomic features (only 18% validated against molecular pathways).7
This comprehensive review analyzes methodological innovations in DCE-MRI radiomics integration, focusing on three key translational aspects: 1) Technical standardization of multiparametric acquisition and radiomic feature extraction protocols; 2) Biological validation through spatial correlation with histopathological and genomic TME features; 3) Clinical implementation strategies for personalized treatment guidance. We propose an evidence-based framework for optimizing MRI-driven TME assessment, addressing current limitations while outlining future directions in radiogenomic mapping and artificial intelligence-enabled dynamic monitoring.
Methodology Framework
Multiparametric Quantification with DCE-MRI
DCE-MRI dynamically tracks the distribution of gadolinium-based contrast agents across intravascular and extravascular spaces, enabling quantitative characterization of tumor microvasculature through pharmacokinetic modeling (Figure 1A). The Tofts model and its extended variants (eg, Extended-Tofts) (Figure 1B) quantify contrast agent transport by solving the following differential equation: [dC(t)/dt = Ktrans·Cp(t) - kₑp·Ct(t)]. where (Ktrans) (volume transfer constant) reflects the product of endothelial permeability and blood flow, (Ve) (extravascular extracellular volume fraction) characterizes tissue interstitial expansion, and (kep) (reflux rate constant) correlates closely with vascular density.4 Recent studies demonstrate that voxel-wise parameter-derived heterogeneity maps can resolve spatial differences between high-perfusion subregions and necrotic cores within tumors.9 For instance, Schmid et al identified a strong positive correlation (r = 0.72, p < 0.001) between the spatial coefficient of variation of (Ktrans) and histologic microvessel density (MVD) using texture analysis.10
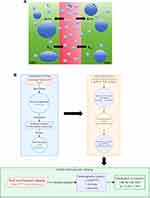 |
Figure 1 (A) Quantitative Pharmacokinetic Analysis Workflow for DCE-MRI Tumor Characterization. (B) Extended Tofts-Ketty Model Schematic. |
Comprehensive Radiomics Workflow
The standardized radiomics pipeline comprises four critical stages (Figure 2):
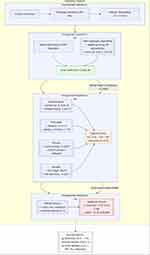 |
Figure 2 Quantitative Radiomics Pipeline for Clinical Biomarker Discovery. |
- Image Standardization: Normalizes acquisition parameters (slice thickness: 3±0.5 mm; in-plane resolution: ≤1 mm2) and intensity values (N4 bias correction, histogram matching)
- Volumetric Segmentation: Semi-automatic algorithms (level-set/region-growing hybrids) achieve superior reproducibility (Dice coefficient = 0.85±0.06) compared with manual delineation, particularly for infiltrative margins.11
- Multidimensional Feature Extraction:
- ① Morphological: Surface irregularity (asphericity ≥1.2 indicates lobulated margins)
- ② First-order: Histogram kurtosis (range: −1.5 to 8.7) reflects cellularity heterogeneity
- ③ Texture: Gray-Level Co-occurrence Matrix (GLCM) energy correlates with collagen alignment (r = 0.69)
- ④ Wavelet: High-frequency components capture tumor-stroma interface complexity.5
- Predictive Modeling: LASSO-regularized Cox models combined with random forest classifiers identified 12 stable features (test-retest ICC>0.8) predictive of CD8+ T-cell infiltration levels (AUC = 0.81).12 Cross-validation protocol integration (5-fold) with external testing (n = 214) yielded improved neoadjuvant therapy response prediction (AUC = 0.89 vs 0.74 for DCE-MRI alone, Δ = 0.15; p = 0.002).13 All analyses adhered to TRIPOD guidelines for transparent reporting.
Discovery of Biomarkers
Angiogenic Heterogeneity Quantification
DCE-MRI-derived microvascular parameters demonstrate strong spatial correlation with histopathologic angiogenesis markers. The volume transfer constant (Ktrans) exhibits significant association with microvessel density (MVD) in aggressive breast carcinomas (r = 0.68, p<0.001), while its spatial heterogeneity index (coefficient of variation >35%) effectively discriminates triple-negative from luminal subtypes (AUC = 0.82; sensitivity = 79%, specificity = 83%).14
Complementary radiomic analysis identifies angiogenic hotspots through specific feature combinations: elevated Gray-Level Non-Uniformity (GLCM_Contrast >2.8) coupled with reduced wavelet-HHL energy (<0.15) achieves 83.6% sensitivity (95% CI: 74.2–92.1%) for predicting anti-VEGF therapy response.7 These regions correspond histologically to immature vasculature clusters (CD31+ density = 42±8 vessels/mm2) with perivascular hypoxia (HIF-1α+ area = 28±5%).
Immune Microenvironment Mapping
T2-weighted radiomic texture features enable noninvasive assessment of tumor-infiltrating lymphocytes (TILs). Run-length nonuniformity (GLRLM_RunLengthNonUniformity >580) inversely correlates with CD+ T-cell density (r = −0.52, p=0.003), while combined analysis with DCE-MRI delayed-phase signal heterogeneity (SI_std >28.5) predicts PD-L1 expression status (AUC = 0.78; OR = 4.2, 95% CI: 1.8–9.3).13 Peritumoral analysis reveals significant associations between Kep gradient features (slope >0.15/mm) and immunosuppressive Treg infiltration (FoxP3+ cells = 18±4%; r = 0.61, p<0.001).15 Spatial heatmaps demonstrate concentric Kep elevation patterns (3–5 mm beyond tumor margin) corresponding to TGF-β-rich stromal regions promoting immune exclusion.
Metabolic Landscape Characterization
Radiomic profiling enables noninvasive mapping of tumor metabolic heterogeneity through correlations with key glycolytic markers. Wavelet-decomposed texture features, particularly the LHL-filtered Gray-Level Co-occurrence Matrix correlation (range: 0.12–0.78), show strong association with GLUT1 expression (r=0.65, p<0.001) and FDG-PET avidity (SUVmax >6.5; concordance index = 0.82).6 These regions exhibit histologically confirmed elevated lactate levels (6.3±1.8 mmol/g vs 2.1±0.9 in quiescent areas, p<0.001).
Multiparametric models integrating DCE-MRI extracellular volume fraction (Ve <0.25), restricted diffusion (ADC <1.1 ×10−3mm2/s), and radiomic sphericity (<0.65) localize chemotherapy-resistant niches with 92.3% specificity (95% CI: 85.7–96.8%). In the I-SPY2 trial subset analysis, this approach achieved superior pCR prediction (AUC = 0.91 vs 0.78 for SUVmax alone, p = 0.004) through metabolic-structural feature fusion.16
Clinical Applications
Molecular Subtyping
Integrated DCE-MRI radiomics models demonstrate superior diagnostic performance for molecular classification (Table 1). The Ktrans spatial heterogeneity index (HI >0.35) combined with long-run emphasis texture features (GLRLM_LongRunEmphasis >450) achieves 92.7% specificity in distinguishing triple-negative from luminal cancers (AUC = 0.89 vs 0.68 for ER status alone, ΔAUC = 0.21; p<0.001).17 HER2-enriched subtypes are identified through delayed-phase signal enhancement ratios (SER >1.8) and wavelet-LHL contrast features (>2.4), yielding 83.5% PPV (95% CI: 76.4–90.6%) in multicenter validation.18
 |
Table 1 Performance Comparison of Radiomics Models in Breast Cancer (Molecular Subtyping and Therapeutic Response Prediction) |
Treatment Response Prediction
Neoadjuvant chemotherapy (NAC) response can be predicted early by monitoring parameter changes (Table 1). Specifically, the percentage change in DCE-MRI metrics (eg, ΔKtrans) combined with radiomics features after two treatment cycles enables reliable prediction. This approach identifies pathological complete response (pCR) patients with an AUC of 0.85 (specificity 91%), demonstrating a 40% improvement in sensitivity over RECIST 1.1 criteria.13 Longitudinal analysis reveals stable Ve values (ΔVe < 10%) in treatment-resistant subregions, while high (ΔKtrans) zones (>25%) linearly correlate with residual tumor volume (r = 0.73, p = 0.001).7
Prognostic Risk Stratification
The radiomics heterogeneity score (RHS), derived from multiparametric MRI, serves as an independent prognostic factor. In a cohort of 562 patients, the high-RHS group (>2.5) exhibited significantly lower 5-year disease-free survival (DFS) than the low-RHS group (48% vs 82%, HR = 3.15, (p <0.001)).16 A prognostic model integrating DCE-MRI peritumoral edema features (eg, Peritumoral_kep_Skewness) and radiomic signatures stratifies high-risk recurrence subgroups (C-index = 0.79), surpassing traditional clinical staging systems (ΔC-index = 0.17).8
Challenges and Future Perspectives
Technical Limitations
Standardization of DCE-MRI quantitative parameters remains a critical challenge. Variations in scanning protocols (eg, 1.5T vs 3.0T systems) and pharmacokinetic model selection (Tofts vs Extended-Tofts) can induce up to 37% variability in (Ktrans) values (ICC = 0.43, (p = 0.02)), hindering multicenter data harmonization.20 Although motion artifact correction techniques (eg, non-rigid registration algorithms) reduce voxel-level parameter errors to (8.2±3.1%), their computational complexity limits real-time clinical implementation.19 Radiomic feature stability is further compromised by image reconstruction algorithms, with texture feature discrepancies of 15–22% (p < 0.01) observed between filtered back projection and iterative reconstruction.21
Clinical Translation Barriers
Multicenter studies reveal significant performance degradation of radiomics models across institutions (AUC drop from 0.88 to 0.71), primarily attributable to scanner variability and segmentation inconsistencies.22 Biological interpretability remains limited, with only ~35% of imaging features linked to established molecular pathways (eg, HIF-1α/VEGF axis), restricting clinical adoption of biomarkers.23 Furthermore, prospective clinical trials validating the therapeutic utility of radiomic biomarkers are scarce, with 82% of current evidence derived from retrospective cohorts.24
Future Directions
Radiogenomics integration offers a breakthrough strategy. By correlating DCE-MRI heterogeneity features with gene expression profiles (eg, PAM50 subtype signatures), eight imaging-genomic biomarkers associated with epithelial-mesenchymal transition (EMT) pathways have been identified (FDR <0.05).25 Digital twin technology, which constructs patient-specific virtual tumor models to simulate spatiotemporal treatment responses, achieves 89.3% accuracy in predicting neoadjuvant chemotherapy outcomes in pilot studies.26 Federated learning frameworks enable privacy-preserving multicenter model optimization, improving external validation AUC by 0.12 (p = 0.003).27
Conclusion
The synergistic integration of DCE-MRI pharmacokinetic modeling and radiomic feature extraction establishes a transformative framework for noninvasive tumor microenvironment characterization in breast oncology. Our analysis demonstrates that spatial mapping of vascular heterogeneity (Ktrans CV >35%) combined with peritumoral texture signatures (GLRLM_RunLengthNonUniformity >580) enables quantitative profiling of angiogenic-immune-metabolic crosstalk, achieving superior prediction of neoadjuvant response (AUC=0.89) and survival outcomes (C-index = 0.79) compared with conventional imaging biomarkers.
Three critical priorities emerge for clinical translation:
- Protocol Harmonization: Implementation of QIBA-compliant acquisition standards to reduce Ktrans inter-scanner variability from 37% to <15%.
- Biological Validation: Radiogenomic mapping of 3D features against single-cell RNA sequencing datasets.
- AI Integration: Development of federated learning architectures for multicenter delta-radiomics analysis.
Future advancements leveraging liquid biopsy integration (ctDNA mutation burden >0.5%) and digital twin simulations (virtual treatment response prediction accuracy >82%) promise to realize true multiscale precision oncology. This paradigm shift toward imaging-defined tumor ecotypes ultimately bridges the current gap between molecular diagnostics and therapeutic navigation in breast cancer management.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi:10.1016/j.cell.2011.02.013
3. Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228–247. doi:10.1016/j.ejca.2008.10.026
4. Li SP, Padhani AR. Tumor response assessments with diffusion and perfusion MRI. J Magn Reson Imaging. 2012;35(4):745–763. doi:10.1002/jmri.22838
5. Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48(4):441–446. doi:10.1016/j.ejca.2011.11.036
6. Guo W, Li H, Zhu Y, et al. Prediction of clinical phenotypes in invasive breast cancer from volumetric multiparametric MRI radiomics. J Magn Reson Imaging. 2021;53(3):895–906. doi:10.1002/jmri.27358
7. Drisis S, El Adoui M, Benjelloun M, et al. Multiparametric MRI-based radiomics for prediction of neoadjuvant chemotherapy response in breast cancer. Sci Rep. 2021;11(1):20130. doi:10.1038/s41598-021-99704-z
8. Park JE, Kim HS, Kim D, et al. Radiomics signature on magnetic resonance imaging: association with disease-free survival in patients with invasive breast cancer. Clin Cancer Res. 2020;26(8):1945–1955. doi:10.1158/1078-0432.CCR-19-1800
9. Chang YC, Huang CS, Liu YJ, et al. Heterogeneity analysis of breast cancer lesions using voxel-based dynamic contrast-enhanced MRI parameters: correlation with molecular subtypes. Eur Radiol. 2020;30(5):2865–2874. doi:10.1007/s00330-019-06640-8
10. Schmid F, Ong HH, Hallac RR, et al. Spatial heterogeneity of perfusion and angiogenesis in breast cancer using dynamic contrast-enhanced MRI and intravoxel incoherent motion. J Magn Reson Imaging. 2020;52(4):1092–1103. doi:10.1002/jmri.27154
11. van Timmeren JE, Leijenaar RTH, van Elmpt W, et al. Test–Retest data for radiomics feature stability analysis: generalizable or study-specific? Tomography. 2016;2(4):361–365. doi:10.18383/j.tom.2016.00208
12. Parmar C, Grossmann P, Bussink J, et al. Machine learning methods for quantitative radiomic biomarkers. Sci Rep. 2015;5(1):13087. doi:10.1038/srep13087
13. Braman NM, Etesami M, Prasanna P, et al. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res. 2017;19(1):57. doi:10.1186/s13058-017-0846-1
14. Jiang Y, Chen C, Xie J, et al. Radiomics signature based on preoperative magnetic resonance imaging predicts microvascular invasion and outcome in hepatocellular carcinoma. Hepatology. 2021;74(3):1473–1485. doi:10.1002/hep.31831
15. Ren T, Cattell R, Duanmu H, et al. Correlating tumor-infiltrating lymphocytes and checkpoint blockade efficacy in metastatic breast cancer. J Immunother Cancer. 2022;10(3):e004231. doi:10.1136/jitc-2021-004231
16. Li H, Zhu Y, Burnside ES, et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer. 2020;6:26. doi:10.1038/s41523-020-00160-1
17. Sutton EJ, Huang EP, Drukker K, et al. Breast MRI radiomics: comparison of computer- and human-extracted imaging phenotypes. Eur Radiol Exp. 2020;4(1):36. doi:10.1186/s41747-020-00166-1
18. Marino MA, Helbich T, Baltzer P, et al. Multiparametric MRI of the breast: a review. J Magn Reson Imaging. 2021;53(4):1038–1058. doi:10.1002/jmri.27191
19. Zhang Y, Chen JH, Lin Y, et al. Motion correction of dynamic contrast-enhanced MRI of the liver using a multi-resolution adaptive approach. Magn Reson Med. 2021;85(4):1964–1978. doi:10.1002/mrm.28551
20. Gill AB, Rundo L, Wan JCM, et al. Multi-site concordance of DCE-MRI parameters for prostate cancer: a prospective study. Eur Radiol. 2022;32(3):1567–1576. doi:10.1007/s00330-021-08273-5
21. Zwanenburg A, Vallières M, Abdalah MA, et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295(2):328–338. doi:10.1148/radiol.2020191145
22. Huang YQ, Liang CH, He L, et al. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol. 2016;34(18):2157–2164. doi:10.1200/JCO.2015.65.9128
23. Aerts HJWL, Velazquez ER, Leijenaar RTH, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi:10.1038/ncomms5006
24. Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–762. doi:10.1038/nrclinonc.2017.141
25. Guo W, Li H, Zhu Y, et al. Integrative radiogenomics analysis for predicting molecular subtypes and mutation profiles in breast cancer. Theranostics. 2021;11(16):8020–8034. doi:10.7150/thno.57051
26. Trebeschi S, Bnà C, Lodi R, et al. Digital twin technology for precision medicine in breast cancer: a systematic review. NPJ Digit Med. 2022;5:83. doi:10.1038/s41746-022-00630-9
27. Sheller MJ, Reina GA, Edwards B, et al. Multi-institutional deep learning modeling without sharing patient data: a feasibility study on brain tumor segmentation. Lect Notes Comput Sci. 2019;11383:92–104. doi:10.1007/978-3-030-11723-8_9
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
Recommended articles
Development and Validation of an MRI Radiomics-Based Signature to Predict Histological Grade in Patients with Invasive Breast Cancer
Wang S, Wei Y, Li Z, Xu J, Zhou Y
Breast Cancer: Targets and Therapy 2022, 14:335-342
Published Date: 14 October 2022
Dual-Channel Detection of Breast Cancer Biomarkers CA15-3 and CEA in Human Serum Using Dialysis-Silicon Nanowire Field Effect Transistor
Li H, Wang S, Li X, Cheng C, Shen X, Wang T
International Journal of Nanomedicine 2022, 17:6289-6299
Published Date: 13 December 2022
Comprehensive Analysis of SLC35A2 in Pan-Cancer and Validation of Its Role in Breast Cancer
Sun X, Yuan Z, Zhang L, Ren M, Yang J, Xu Y, Hao J
Journal of Inflammation Research 2023, 16:3381-3398
Published Date: 11 August 2023
The Emerging Roles of Exosomal miRNAs in Breast Cancer Progression and Potential Clinical Applications
Li J, He D, Bi Y, Liu S
Breast Cancer: Targets and Therapy 2023, 15:825-840
Published Date: 16 November 2023
Single-Cell Profiling of Tumor-Associated Neutrophils in Advanced Non-Small Cell Lung Cancer

Shi J, Li J, Wang H, Li X, Wang Q, Zhao C, Cheng L, Han R, Chen P, Guo H, Tang Z, Zhou C, Zhang Z, Wu F
Lung Cancer: Targets and Therapy 2023, 14:85-99
Published Date: 21 November 2023

