Back to Journals » Journal of Inflammation Research » Volume 17
Identification of Diagnostically Relevant Biomarkers in Patients with Coronary Artery Disease by Comprehensive Analysis
Authors Wu Z, Mo S, Huang Z, Zheng B
Received 3 September 2024
Accepted for publication 28 November 2024
Published 5 December 2024 Volume 2024:17 Pages 10495—10513
DOI https://doi.org/10.2147/JIR.S494438
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tara Strutt
Zimin Wu,1,* Sisi Mo,2,* Zuyuan Huang,1 Baoshi Zheng1
1Department of Cardiovascular Surgery Ward, The First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi Zhuang Autonomous Region, 530021, People’s Republic of China; 2Department of Medical Research, The First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi Zhuang Autonomous Region, 530021, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Baoshi Zheng, Department of Cardiovascular Surgery Ward, the First Affiliated Hospital of Guangxi Medical University, No. 6 Shuangyong Road, Qingxiu District, Nanning, Guangxi Zhuang Autonomous Region, 530021, People’s Republic of China, Tel +86 0771-5355280, Email [email protected]
Background: Peripheral biomarkers are becoming an important method by which to monitor the progression of coronary artery disease (CAD). Not only are they minimally invasive and early detection, but they can also be used for classification and diagnosis of disease as well as prognostic assessment. Currently, this approach is still in the exploratory stage. The purpose of this research is to determine the diagnostic value and therapeutic potential of the endoplasmic reticulum stress (ERS) genes in CAD.
Methods: The clinical information and RNA sequence data were obtained from the GEO database and subsequently subjected to a series of optimization and visualization processes using various analytical techniques, including WGCNA, LASSO, SVM-RFE feature selection, random forest (RF), and XGBoost, as well as R software and Cytoscape. Finally, immunofluorescence was used to validate the analysis.
Results: We identify 6 key ERS differentially expressed genes (ERS-DEGs) (UFL1, HSPA1A, ERLIN1, LRRK2, ERN1, SERINC3) for constructing diagnostic models. They showed qualified diagnostic ability as biomarkers of CAD within training dataset (AUC = 0.803) and validation dataset (AUC = 0.776 and 0.797). Association analyses showed that peripheral immune cells, immune checkpoint genes and Human Leukocyte Antigen (HLA) genes had characteristic distributions in CAD and were closely related to specific ERS genes. Meanwhile, we found that HSPA1A may involve the MAPK signaling pathway in CAD.
Conclusion: We constructed an efficient diagnostic model based on 6 key ERS-DEGs and explored their regulatory networks and effects on the CAD immune microenvironment. UFL1, HSPA1A, ERLIN1, LRRK2, ERN1, SERINC3 are expected to be biomarkers for CAD.
Keywords: endoplasmic reticulum stress, coronary artery disease, HSPA1A, immune microenvironment
Introduction
Coronary artery disease (CAD) is defined as myocardial dysfunction and/or organic lesions due to coronary artery stenosis and inadequate blood supply, mostly caused by risk factors such as smoking, obesity, malnutrition, hypertension, hyperlipidemia and diabetes. It is the foremost cause of death globally and places a heavy burden on societies and individuals.1–3 In addition, the immune response is associated with CAD.4 Molecular targeted therapy is a promising research direction in precision medicine. For this reason, it is crucial to uncover biomarkers for prognostic analysis of CAD.
Endoplasmic reticulum (ER) consists of biological membranes involved in protein synthesis, modification, and transport in eukaryotic cells and regulates lipid and steroid metabolism as well as calcium levels.5–7 Oxidative stress, ischemic injury, disturbances in calcium homeostasis and enhancement of normal and/or defectively folded proteins can lead to unfolded proteins accumulating in the ER, resulting in ER dysfunction, which is called endoplasmic reticulum stress (ERS).8 ERS triggers the unfolded protein response (UPR) to maintain ER homeostasis.9 The UPR has three classical pathways, inositol-requiring enzyme-1α (IRE1α), protein kinase RNA like ER kinase (PERK) and activating transcription factor 6 (ATF6), which reduce unfolded proteins through increasing ER-binding proteins, inhibiting protein biosynthesis and accelerating unfolded protein degradation.10,11 UPR is an adaptive response, but persistent or excessive activation of UPR initiates cell death through multiple pathways.12 When UPR is over-activated, three transmembrane proteins induce autophagy through IRE1α/TRAF2/JNK/Beclin1 signaling pathway, PERK/ATF4/ATG12/CHOP signaling pathway and ATF6/CHOP signaling pathway. In addition, UPR activates the IRE1α/TRAF2, IRE1α/XBP1/CHOP, PERK/ATF4/CHOP, ATF6/CHOP signaling pathway, which play a key role in the association between ERS and apoptosis. It also mediates apoptosis by degrading certain mRNA. Furthermore, UPR is mainly induced ferroptosis through PERK/SLC7A11 and IRE1α/JNK pathways, involved in pyroptosis through activation of NLRP3, and induced necroptosis through RIPK1/RIPK3/MLKL and RIPK3/CaMKKII signaling pathways.13,14 These extensive cellular processes, triggered by ERS initiation of UPR, disrupt ER homeostasis and thus contribute to the threshold and evolvement of cardiovascular disease.8 Therefore, ER is now considered to be an important organelle that determines cell survival and death. Growing evidence shows that ERS is associated with CAD. Chen J et al found that SERCA2a attenuates myocardial ischemia-reperfusion injury through inhibition of ERS.15 Liu Z et al showed that advanced glycation end products exacerbate coronary microvascular dysfunction by activating ERS-mediated PERK/CaN/NFATc4 signaling in Cardiac microcirculation endothelial cells.16 However, the potential impact of ERS on CAD requires a more comprehensive analysis.
In this context, one of the crucial tools to analyze the role of ERS in CAD is through bioinformatics. We screened and optimized ERS genes related to CAD, developed a promising diagnostic prediction model and validated it in vitro.
Materials and Methods
Data Acquisition
The following datasets were downloaded from the GEO (http://www.ncbi.nlm.nih.gov/geo/): GSE20681: 198 human blood samples were collected as the training dataset, which included 99 CAD patients and 99 healthy controls. GSE20680: 195 human blood samples with 143 CAD patients and 52 healthy controls were used as the validation dataset. GES42148:24 human blood samples with 13 CAD patients and 11 healthy controls were used as the validation dataset.
Acquisition and Optimization of Differential ERS Genes
Screening for Significantly Different ERS Genes
ERS genes were obtained by MSigDB of the GSEA (http://software.broadinstitute.org/gsea/downloads.jsp).17 The corresponding genes were extracted from the GSE20681 and were divided into CAD and Control (CTRL) groups. The ERS differentially expressed genes (ERS-DEGs) was analyzed using the R software “limma”18 and “cor” was used for correlation calculations.
WGCNA Screens Modules for Significant Correlation of Disease States
We used “WGCNA” of R software to screen for modules that were significantly associated with the phenotypic grouping of the samples (the set of modules contained a minimum of 150 genes and cutHeight = 0.995).19 We selected the genes highly associated with the phenotype in GSE20681 and compared with the ERS-DEGs, keeping the overlapping part. The “clusterProfiler” of R software performed enrichment analysis of GO and KEGG on the screened significantly ERS-DEGs (FDR < 0.05).20 Interaction proteins with overlapping ERS-DEGs were obtained from the STRING (http://string-db.org/) and made visualization via “Cytoscape”.21,22
Construction of a Diagnostic Model
One-way logistic regression analyses of ERS-DEGs in GSE20681 were conducted using “rms” in R software.23 The LASSO algorithm, SVM-RFE feature selection, random forest (RF) and XGBoost algorithms were employed in conjunction to identify the most optimal ERS-DEGs.24–26 Kruskal–Wallis was utilized to assess the risk values of CAD and CTRL samples. A diagnostic model based on ERS-DEGs was developed in GSE20681 using a support vector machine approach.27 The reliability of the diagnostic model was assessed by receiver operating characteristic (ROC) curve.28
Construction of Nomogram
The “rms” in R software was used to construct the nomogram and draw the correction folds for the ERS-DEGs.29 The C-index coefficients were calculated using “survcomp”.30 In addition, we plotted decision curves for each of the ERS-DEGs using “rmda” to see the benefit of each factor on survival.31
Diagnostic ERS-DEGs and Immune Correlation Analysis
Evaluation of Immune Cell Proportions
We analyzed the distribution of immune cells in GSE20681 using CIBERSORT and Kruskal–Wallis tests.32 We then conducted correlation analyses using the R software “cor”.
Immune Checkpoint Gene and Human Leukocyte Antigen Gene Correlation Analysis
Immune checkpoint genes and Human Leukocyte Antigen (HLA) family genes were extracted from GSE20681 and correlations with diagnostic ERS-DEGs were calculated by “cor” in R software.
Constructing a KEGG Pathway- ERS-DEGs-Immune Landscape Network
The KEGG pathway-ERS-DEGs-immune landscape network was constructed based on immune cells, immune checkpoint genes, HLA genes and signaling pathways associated with ERS. The network is presented via Cytoscape.22
Immunofluorescence
H9C2 Rat cardiomyocytes were purchased from The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences (Shanghai, China), cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin (Gibco; Thermo Fisher Scientific, Inc.), at 37 °C under 5% CO2. We set up two groups: Control (Con) and hypoxia/reoxygenation (H/R) and intervention after cell adhesion (hypoxia 8h, reoxygenation 12h). Immunofluorescence was performed according to the previous method.33 Antibodies: HSPA1A (1:100), GRP78 (1:200), ATF6 (1:100), PERK (1:100), IRE1α (1:100), p38 MAPK (1:50), p-p38 MAPK (1:50), JNK (1:200), p-JNK (1:200), ERK (1:200). The relative fluorescence intensity was calculated via ImageJ.
Results
Screening for ERS-DEGs
We extracted 295 ERS-related genes from GSE20681 and calculated the differences between CAD and CTRL groups. Next, we screened 22 ERS-DEGs. HSPA1A, ERO1A, TRIM25, CEBPB, LRRK2, NRBF2, ERLIN1, CASP4, ERN1, MAP3K5, TMEM33, ALOX5, HERPUD1, TMBIM6, SERINC3, EXTL3,CDK5RAP3 were up-regulated (P < 0.05), while RNF5, UFL1, YIF1A, TP53, GET4 were down-regulated (P < 0.05). Some of them showed obvious positive correlations, as for HSPA1A/CEBPB (r = 0.705) and CASP4/LRRK2 (r = 0.643). The other parts showed negative correlations, such as YIF1A/LRRK2 (r = −0.551) and UFL1/TRIM25 (r = −0.419) (Figure 1A–B). The WGCNA was performed in GSE20681 and the value of the power was 8 (Figure 2A). We categorize the ERS-DEGs into eight modules and select two with correlation absolute values higher than 0.3 for a total of 638 genes (Figure 2B–C). We then compared some of these with 22 ERS-DEGs and obtained 10 overlapping ERS-DEGs (ALOX5, CEBPB, ERLIN1, ERN1, HSPA1A, LRRK2, NRBF2, SERINC3, TRIM25, UFL1). Next, we performed GO and KEGG analysis for 10 ERS-DEGs and verified that these genes are associated with ERS, protein processing and autophagy (Figure 2D–E). We finally constructed 10 ERS-DEGs product protein interactions networks and retained connections with linkage coefficients higher than 0.4. We found that HSPA1A was in the core (Figure 2F).
 |
Figure 1 ERS-DEGs identification. (A) Distribution of significantly ERS-DEGs; (B) Heatmap of the correlation of significantly ERS-DEGs. |
 |
Figure 2 Continued. |
Construction of a Diagnostic Model
Single factor logistic regression demonstrated that P < 0.05 for all 10 ERS-DEGs in GSE20681 (Figure 3A). We subsequently applied four algorithms—LASSO (Figure 3B), SVM-RFE (Figure 3C), RF (Figure 3D), and XGBoost (Figure 3E)—to identify relevant features. The features selected by these methods were intersected using Venn analysis (Figure 3F), leading to the identification of 6 key ERS-DEGs (UFL1, HSPA1A, ERLIN1, LRRK2, ERN1, and SERINC3) for constructing a diagnostic model. We compared risk score in disease and control in GSE20681, GSE20680 and GSE42148 (Figure 4A) and the distribution of the 6 key ERS-DEGs (Figure 4C). Diagnostic efficacy of the 6 key ERS-DEGs was assessed by ROC curves (Figure 4B) for the training dataset (AUC = 0.803), validation dataset (AUC = 0.776 and 0.797). The results demonstrate that these 6 key ERS-DEGs exhibit favourable diagnostic potential.
Figure 3 Continued.
Construction of the Nomogram
We constructed nomogram with 6 key ERS-DEGs in GSE20681 and GSE20680. Each gene was projected upward to a point and the sum of the variable scores was translated into the disease risk of the patient. A high total score corresponds to a high risk of disease (Figure 5A). The calibration curve (Figure 5B), decision curve analysis (DCA) and clinical impact curve (CIC) reveal the significant ability of nomograms to predict in CAD (Figure 5C).
 |
Figure 5 The nomogram, DCA and CIC. (A) Nomogram based on the joint diagnosis of 6 key ERS-DEGs; (B) calibration curve; (C) DCA (up) and CIC (down). |
Immunity Assessment and Construction of KEGG Pathway-6 Key ERS-DEGs -Immunity Landscape Network
Considering the importance of multiple immune components in CAD, we screened 5 immune cell types (B cell naive, CD8+ T cell, NK cell activated, Eosinophil, Neutrophil) with markedly different distributions in the CTRL and CAD groups (Figure 6A). Next, we explored the intrinsic linkage of immune cells in CAD to the 6 key ERS-DEGs. The expression of ERLIN1, HSPA1A, LRRK2, SERINC3 were significantly upregulated in neutrophils, with LRRK2 being the most significant (P < 0.005), while UFL1 was significantly downregulated in neutrophils (P < 0.005). The same trend was observed for ERLIN1, HSPA1A, SERINC3 and UFL1 in NK cells, but LRRK2 was not significantly correlated with NK cells. Interestingly, ERLIN1, HSPA1A, LRRK2 and SERINC3 were down-regulated in CD8+ T cell, with LRRK2 being the most significant (P < 0.005) and UFL1 being their opposite. (Figure 6B). To find the potential of the 6 key ERS-DEGs in immunotherapy and gene therapy, we correlated the immune checkpoint genes and HLA family genes in GSE20681 with 6 key ERS-DEGs. The results indicated that ERN1 was significantly up-regulated in CTLA4, CD86 and TIGIT, with the strongest association with CRLA4 (P < 0.005), and showed significant down-regulation in CD27 (P < 0.005). HSPA1A was significantly up-regulated in CD274 (P < 0.005) and down-regulated in CD40 and ICOS (P < 0.005). LRRK2 was associated with most of the immune checkpoint genes except CD86 and IDO1, most of which showed a trend of down-regulation (P<0.005). UFL1 was significantly up-regulated in CD40, TIGIT and ICOS in addition to being down-regulated in CD27 (P < 0.005). While SERINC3 and ERLIN1 were associated with a few immune checkpoint genes (Figure 6C). In HLA gene association analysis, HSPA1A was upregulated with all currently known HLA-I molecules (HLA-A, HLA-B, HLA-C, HLA-E, HLA-F, HLA-G) and some HLA-II molecules (HLA-DMB, TAP1) (P < 0.005), with the strongest positive correlation with TAP1 (P < 0.005). SERINC3 was up-regulated only in HLA-G (P < 0.01), down-regulated in the rest of HLA genes and significant in HLA-DMA (P < 0.005) (Figure 6D). To screen for pathways that are potentially participating in immune regulation, we recombined the immune information obtained above that was notably correlated with the 6 key ERS-DEGs, and constructed the KEGG pathway-6 key ERS-DEGs-immunity landscape network. In this network, we found that HSPA1A was associated with the MAPK signaling pathway (Figure 6E).
Figure 6 Continued.
Expression of ERS-Related Proteins and MAPK Pathway Proteins in H9C2
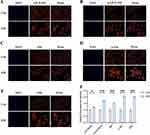
H/R H9C2 cardiomyocytes were used to simulate the pathogenesis of CAD to investigate whether ERS occurred. The results showed that the relative fluorescence intensity of GRP78, the companion molecule of ER and master of the UPR, was enhanced in cardiomyocytes after hypoxia reoxygenation. The same is true for the three downstream ER proteins of UPR: PERK, ATF6 and IRE1α (Figure 7). Next, we explored the possibility of HSPA1A contributing to the simulated CAD via the MAPK pathway. The relative fluorescence intensity of HSPA1A was higher in H/R group than that in Con group (Figure 8). The same happened to the three key proteins in MAPK pathway-p38 MAPK, JNK and ERK. Notably, p-p38 MAPK and p-JNK were also upregulated in the H/R group (Figure 9).
Discussion
ERS is both a cause and consequence of CAD, imposing significant stress on the ER environment. This leads to the accumulation of misfolded proteins and disruption of ER homeostasis, which are closely linked to cardiovascular dysfunction.8 Our study identified significant expression of key proteins from the three classical UPR pathways in H/R-treated H9C2 cardiomyocytes. Based on these findings, 6 key ERS-DEGs (UFL1, HSPA1A, ERLIN1, LRRK2, ERN1, SERINC3) were identified as potential diagnostic markers for CAD. Additionally, an immune landscape network and a nomogram were developed for clinical application. HSPA1A, validated through immunofluorescence, is speculated to influence CAD progression via the MAPK pathway.
Heat shock protein family A member 1A (HSPA1A), a key member of the HSP70 family encoded by the HSPA gene, is often referred to as HSP70 due to its representative function.34 Widely recognized as a stress response protein, HSPA1A is induced by CAD-related stresses, including oxidative, hemodynamic, and inflammatory stress. Its elevated expression and synthesis in the heart under prolonged hypervolemic load were identified as early as 30 years ago.35 Consistent with previous findings,36–38 our study observed increased serum levels of HSPA1A in CAD patients. Evidence suggests that HSPA1A participates in CAD pathology through the MAPK pathway, as indicated by enhanced fluorescence intensity of HSPA1A and MAPK pathway proteins in H/R cardiomyocytes. Additionally, HSPA1A inhibits ABCA1 and ABCG1 via the JNK/Elk-1 pathway, while the SIRT1/HSF1/HSP molecular axis promotes arterial lipid deposition and atherosclerosis progression.39,40 HSPA1A also activates monocyte TLR4 signaling, promoting cardiac inflammation,41 and enhances vascular smooth muscle cell proliferation, collagen secretion, and fibronectin release via AP-1.42 While some studies indicate its anti-atherosclerotic effects,43,44 functional differences across the ecological niche may explain its controversial role in CAD.42 Further research is needed to elucidate these mechanisms.
Endoplasmic reticulum to nucleus signaling 1 (ERN1, also as IRE1α), a critical UPR stress sensor, was found to be elevated in the peripheral blood of CAD patients. In H/R cardiomyocytes, ERN1 increased JNK phosphorylation and activated XBP1, regulating pathological precursors of atherosclerosis, including hyperlipidemia, insulin resistance, and angiogenesis.45 ERN1 promotes ox-LDL uptake and foam cell formation by upregulating PPAR-γ and CD36 through XBP1.46 It also mediates T cell differentiation, with CD8+ T cell accumulation potentially exacerbating atherosclerosis.47,48 Additionally, IRE1α/XBP1 signaling drives dendritic cell lipid efferocytosis and foam cell formation,49,50 while pro-inflammatory pathways (IRE1α/XBP1s/NF-κB and HIF1α) increase secretion of IL-8, IL-1β, IL-6, TNF-α, and MCP-1.46 Beyond UPR signaling, ERN1 contributes to coronary endothelial dysfunction via the IRE1α-JNK-c-Jun/AP-1-sEH pathway.51
Leucine rich repeat kinase 2 (LRRK2), implicated in central nervous system and immune disorders,52,53 was elevated in myocardial infarction periphery. LRRK2-deficient mice showed reduced mortality, infarct size, ventricular fibrosis, and inflammation post-infarction.54 LRRK2 facilitates neutrophil chemotaxis,55 enabling LDL cholesterol extravasation and plaque destabilization via cytokine and adhesion factor release.56 It also enhances caspase-3 activity, increasing apoptotic cardiomyocytes and inflammatory cytokines (TNF-α, IL-1, IL-6) in CAD.54
ER lipid raft associated 1 (ERLIN1) and UFM1 specific ligase 1 (UFL1) are associated primarily with CAD. ERLIN1, a regulator of sterol regulatory element-binding proteins, is involved in IP3R processing and lipid metabolism.57 Its interaction with AMBRA1 in the MAM raft domain facilitates cholesterol transport and autophagosome formation, relevant to CAD pathogenesis.58–60 UFL1, a Ufm1 E3 ligase, protects cardiomyocytes from ERS-induced apoptosis through the PERK pathway, although its role in NF-κB and calcium regulation remains unproven.61
Serine incorporator 3 (SERINC3) was identified for the first time in this study. While its specific role in CAD remains unclear, its family members are implicated in cholesterol metabolism,62 suggesting a similar mechanism for SERINC3.
These 6 key ERS-DEGs play critical roles in CAD pathogenesis, including oxidative stress, inflammation, ER dysfunction, metabolic abnormalities, and apoptosis. Their aberrant expression represents promising biomarkers for early diagnosis and disease severity assessment. Peripheral blood biomarker detection offers a simpler, non-invasive alternative suitable for community-level risk screening and progression monitoring. Our ERS diagnostic model, based on these six genes, demonstrated strong diagnostic performance (training set, AUC = 0.803; validation sets, AUC = 0.776 and 0.797). HSPA1A, particularly, has been proposed as an early-stage CAD biomarker,63,64 with levels positively correlating with disease severity.38 Additionally, the UFL1-FHL5 locus is a leading candidate gene for CAD/myocardial infarction risk.65
Although these 6 key ERS-DEGs were linked to immune checkpoints and HLA genes in this study, their CAD-specific roles are yet unreported, though associations with other diseases exist.66–69 Immune checkpoints like PD-L1 and CTLA4 maintain cardiac immune homeostasis,70 while HLA genes contribute to certain heart diseases.71,72 Investigating the interplay between ERS, immune checkpoints, and HLA in CAD holds significant potential for advancing our understanding of the disease.
Building on the potential of ERS-related genes in CAD, HSPA1A emerges as a promising therapeutic target due to its antioxidative, anti-inflammatory, and cytoprotective properties. Allicin has been shown to enhance HSPA1A expression, mitigating atherosclerosis, improving endothelial function, and reducing myocardial injury and thrombosis risk.73 Similarly, 17-AAG stabilizes HSPA1A, significantly reducing cardiomyocyte apoptosis in murine myocardial infarction models.74 In age-related cardiac amyloidosis, HSPA1A inducers such as GGA have demonstrated efficacy in reducing amyloid fibril deposition and alleviating myocardial conduction abnormalities.75,76
In parallel, LRRK2 has emerged as a poor prognostic marker in CAD patients, primarily due to its role in mitochondrial dysfunction and inflammation.54 Targeting LRRK2 has shown promise, with XL01126—a potent, blood-brain barrier-penetrant LRRK2 degrader—demonstrating potential in mitigating inflammation and mitochondrial dysfunction in cardiovascular diseases.77
ERN1 offers a multidimensional therapeutic pathway in CAD. Pharmacological agents such as GLP-1 receptor agonists and SGLT2 inhibitors suppress ERN1 expression, benefiting CAD patients with glucose metabolism abnormalities.78 Experimental drugs like GSK2850163, which selectively inhibit ERN1-related signaling pathways, reduce endothelial dysfunction and oxidative stress.51 Moreover, Ginkgolide K, by activating the ERN1/XBP1 pathway, enhances autophagy and reduces infarct size in myocardial infarction models.79
Although research into SERINC3, ERLIN1, and UFL1 is still at an early stage, their involvement in ERS regulation and CAD pathogenesis is gaining recognition. For example, UFL1 deficiency is associated with cardiac hypertrophy, which can be alleviated by TUDCA, a known ERS inhibitor.80 Further studies are needed to clarify their molecular mechanisms and validate their roles in CAD.
In summary, targeting HSPA1A, LRRK2, and ERN1, along with emerging ERS-related genes like SERINC3, ERLIN1, and UFL1, represents promising therapeutic directions in CAD management. These findings highlight opportunities to mitigate oxidative stress, inflammation, mitochondrial dysfunction, and ERS, offering innovative strategies to enhance cardiovascular outcomes.
This study complements these findings by integrating multi-omics data analysis with experimental validation to construct a diagnostic model for ERS-related genes in CAD. Using publicly available datasets (GSE20681, GSE20680, GSE42148) and techniques such as WGCNA and machine learning algorithms (eg, LASSO, SVM-RFE, RF and XGBoost), the study systematically identified ERS-related genes. Additionally, analyses of immune cell proportions, immune checkpoint genes, and HLA gene correlations provided insights into the immunoregulatory mechanisms underlying CAD. Immunofluorescence experiments further validated the expression of key ERS signaling proteins, underscoring the translational potential of these findings. However, the study is limited by its reliance on publicly available datasets, which may introduce heterogeneity, and the lack of validation in human samples. While the immune-related analyses suggest potential mechanisms, further investigation is required to delineate specific molecular pathways and enhance clinical applicability. Future research should focus on bridging these gaps to advance the scientific and translational potential of these discoveries.
Conclusion
We developed an efficient diagnostic model utilizing CAD-associated online datasets and investigated the regulatory networks of the 6 key ERS-DEGs (UFL1, HSPA1A, ERLIN1, LRRK2, ERN1 and SERINC3), along with their roles in modulating the CAD immune microenvironment. These genes show promising potential as biomarkers for CAD, offering valuable insights for translational research on ERS-DEGs in CAD. Nevertheless, this study remains preliminary and is limited in scope. To refine and validate our findings, large-scale clinical trials and single-cell analyses are necessary, marking the direction of our future work.
Data Sharing Statement
The datasets generated during and/or analyzed during the current study are available from GEO database (http://www.ncbi.nlm.nih.gov/geo/) and corresponding author on reasonable request.
Ethics Statement
The study involving human data was approved by the Ethics Committee of The First Affiliated Hospital of Guangxi Medical University (2024-E761-01).
Acknowledgments
We express our gratitude to the team at the GEO for allowing us to utilize their invaluable data.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the National Natural Science Foundation of China (No. 8206020365).
Disclosure
The authors declare no conflicts of interest regarding the publication of this paper.
References
1. Malakar AK, Choudhury D, Halder B, Paul P, Uddin A, Chakraborty S. A review on coronary artery disease, its risk factors, and therapeutics. J Cell Physiol. 2019;234(10):16812–16823. doi:10.1002/jcp.28350
2. Lee YH, Fang J, Schieb L, Park S, Casper M, Gillespie C. Prevalence and trends of coronary heart disease in the United States, 2011 to 2018. JAMA Cardiol. 2022;7(4):459–462. doi:10.1001/jamacardio.2021.5613
3. Khera AV, Kathiresan S. Genetics of coronary artery disease: discovery, biology and clinical translation. Nat Rev Genet. 2017;18(6):331–344. doi:10.1038/nrg.2016.160
4. Kott KA, Vernon ST, Hansen T, et al. Single-cell immune profiling in coronary artery disease: the role of state-of-the-art immunophenotyping with mass cytometry in the diagnosis of atherosclerosis. J Am Heart Assoc. 2020;9(24):e017759. doi:10.1161/jaha.120.017759
5. Wang M, Kaufman RJ. Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature. 2016;529:7586):326–35. doi:10.1038/nature17041
6. Schwarz DS, Blower MD. The endoplasmic reticulum: structure, function and response to cellular signaling. Cell Mol Life Sci. 2016;73(1):79–94. doi:10.1007/s00018-015-2052-6
7. Oakes SA, Papa FR. The role of endoplasmic reticulum stress in human pathology. Annu Rev Pathol. 2015;10(1):173–194. doi:10.1146/annurev-pathol-012513-104649
8. Ren J, Bi Y, Sowers JR, Hetz C, Zhang Y. Endoplasmic reticulum stress and unfolded protein response in cardiovascular diseases. Nat Rev Cardiol. 2021;18(7):499–521. doi:10.1038/s41569-021-00511-w
9. Hetz C, Zhang K, Kaufman RJ. Mechanisms, regulation and functions of the unfolded protein response. Nat Rev Mol Cell Biol. 2020;21(8):421–438. doi:10.1038/s41580-020-0250-z
10. Bi X, Zhang G, Wang X, et al. Endoplasmic reticulum chaperone GRP78 protects heart from ischemia/reperfusion injury through akt activation. Circ Res. 2018;122(11):1545–1554. doi:10.1161/circresaha.117.312641
11. Kopp MC, Larburu N, Durairaj V, Adams CJ, Ali MMU. UPR proteins IRE1 and PERK switch BiP from chaperone to ER stress sensor. Nat Struct Mol Biol. 2019;26(11):1053–1062. doi:10.1038/s41594-019-0324-9
12. Hetz C, Papa FR. The unfolded protein response and cell fate control. Mol Cell. 2018;69(2):169–181. doi:10.1016/j.molcel.2017.06.017
13. An Y, Wang X, Guan X, et al. Endoplasmic reticulum stress-mediated cell death in cardiovascular disease. Cell Stress Chaperones. 2024;29(1):158–174. doi:10.1016/j.cstres.2023.12.003
14. Ajoolabady A, Wang S, Kroemer G, et al. ER stress in cardiometabolic diseases: from molecular mechanisms to therapeutics. Endocr Rev. 2021;42(6):839–871. doi:10.1210/endrev/bnab006
15. Chen J, Liu Y, Pan D, et al. Estrogen inhibits endoplasmic reticulum stress and ameliorates myocardial ischemia/reperfusion injury in rats by upregulating SERCA2a. Cell Commun Signal. 2022;20(1):38. doi:10.1186/s12964-022-00842-2
16. Liu Z, Zhu H, Ma Y, et al. AGEs exacerbates coronary microvascular dysfunction in NoCAD by activating endoplasmic reticulum stress-mediated PERK signaling pathway. Metabolism. 2021;117:154710. doi:10.1016/j.metabol.2021.154710
17. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi:10.1073/pnas.0506580102
18. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi:10.1093/nar/gkv007
19. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9(1):559. doi:10.1186/1471-2105-9-559
20. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287. doi:10.1089/omi.2011.0118
21. Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–d612. doi:10.1093/nar/gkaa1074
22. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
23. Pan X, Jin X, Wang J, Hu Q, Dai B. Placenta inflammation is closely associated with gestational diabetes mellitus. Am J Transl Res. 2021;13(5):4068–4079.
24. Goeman JJ. L1 penalized estimation in the Cox proportional hazards model. Biom J. 2010;52(1):70–84. doi:10.1002/bimj.200900028
25. Wei C, Wei Y, Cheng J, et al. Identification and verification of diagnostic biomarkers in recurrent pregnancy loss via machine learning algorithm and WGCNA. Front Immunol. 2023;14:1241816. doi:10.3389/fimmu.2023.1241816
26. Shin H. XGBoost regression of the most significant photoplethysmogram features for assessing vascular aging. IEEE J Biomed Health Inform. 2022;26(7):3354–3361. doi:10.1109/jbhi.2022.3151091
27. Arnold PG, Kaya E, Reisert M, et al. Support vector machine-based spontaneous intracranial hypotension detection on brain MRI. Clin Neuroradiol. 2022;32(1):225–230. doi:10.1007/s00062-021-01099-x
28. Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf. 2011;12(1):77. doi:10.1186/1471-2105-12-77
29. Shan S, Chen W, Jia JD. Transcriptome analysis revealed a highly connected gene module associated with cirrhosis to hepatocellular carcinoma development. Front Genet. 2019;10:305. doi:10.3389/fgene.2019.00305
30. Harrell FE Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15(4):361–387. doi:10.1002/(sici)1097-0258(19960229)15:4<361:Aid-sim168>3.0.Co;2-4
31. Mayr A, Schmid M. Boosting the concordance index for survival data--A unified framework to derive and evaluate biomarker combinations. PLoS One. 2014;9(1):e84483. doi:10.1371/journal.pone.0084483
32. Kawada JI, Takeuchi S, Imai H, et al. Immune cell infiltration landscapes in pediatric acute myocarditis analyzed by CIBERSORT. J Cardiol. 2021;77(2):174–178. doi:10.1016/j.jjcc.2020.08.004
33. Li H, Yang DH, Zhang Y, et al. Geniposide suppresses NLRP3 inflammasome-mediated pyroptosis via the AMPK signaling pathway to mitigate myocardial ischemia/reperfusion injury. Chin Med. 2022;17(1):73. doi:10.1186/s13020-022-00616-5
34. Radons J. The human HSP70 family of chaperones: where do we stand? Cell Stress Chaperones. 2016;21(3):379–404. doi:10.1007/s12192-016-0676-6
35. Delcayre C, Samuel JL, Marotte F, Best-Belpomme M, Mercadier JJ, Rappaport L. Synthesis of stress proteins in rat cardiac myocytes 2-4 days after imposition of hemodynamic overload. J Clin Invest. 1988;82(2):460–468. doi:10.1172/jci113619
36. Tsoporis JN, Triantafyllis AS, Kalogeropoulos AS, et al. Differential expression of circulating damage-associated molecular patterns in patients with coronary artery ectasia. Biomolecules. 2023;14(1):10. doi:10.3390/biom14010010
37. Eapen DJ, Manocha P, Patel RS, et al. Aggregate risk score based on markers of inflammation, cell stress, and coagulation is an independent predictor of adverse cardiovascular outcomes. J Am Coll Cardiol. 2013;62(4):329–337. doi:10.1016/j.jacc.2013.03.072
38. Zhang X, Xu Z, Zhou L, et al. Plasma levels of Hsp70 and anti-Hsp70 antibody predict risk of acute coronary syndrome. Cell Stress Chaperones. 2010;15(5):675–686. doi:10.1007/s12192-010-0180-3
39. Zhao ZW, Zhang M, Chen LY, et al. Heat shock protein 70 accelerates atherosclerosis by downregulating the expression of ABCA1 and ABCG1 through the JNK/Elk-1 pathway. Biochim Biophys Acta Mol Cell Biol Lipids. 2018;1863(8):806–822. doi:10.1016/j.bbalip.2018.04.011
40. Bruxel MA, Tavares AMV, Zavarize Neto LD, et al. Chronic whole-body heat treatment relieves atherosclerotic lesions, cardiovascular and metabolic abnormalities, and enhances survival time restoring the anti-inflammatory and anti-senescent heat shock response in mice. Biochimie. 2019;156:33–46. doi:10.1016/j.biochi.2018.09.011
41. Satoh M, Shimoda Y, Akatsu T, Ishikawa Y, Minami Y, Nakamura M. Elevated circulating levels of heat shock protein 70 are related to systemic inflammatory reaction through monocyte Toll signal in patients with heart failure after acute myocardial infarction. Eur J Heart Fail. 2006;8(8):810–815. doi:10.1016/j.ejheart.2006.03.004
42. Costa-Beber LC, Hirsch GE, Heck TG, Ludwig MS. Chaperone duality: the role of extracellular and intracellular HSP70 as a biomarker of endothelial dysfunction in the development of atherosclerosis. Arch Physiol Biochem. 2022;128(4):1016–1023. doi:10.1080/13813455.2020.1745850
43. Poznyak AV, Orekhova VA, Sukhorukov VN, Khotina VA, Popov MA, Orekhov AN. Atheroprotective aspects of heat shock proteins. Int J Mol Sci. 2023;24(14):11750. doi:10.3390/ijms241411750
44. Nagai M, Kaji H. Thermal effect on heat shock protein 70 family to prevent atherosclerotic cardiovascular disease. Biomolecules. 2023;13(5):867. doi:10.3390/biom13050867
45. Wang T, Zhou J, Zhang X, et al. X-box binding protein 1: an adaptor in the pathogenesis of atherosclerosis. Aging Dis. 2023;14(2):350–369. doi:10.14336/ad.2022.0824
46. Zhou ZY, Wu L, Liu YF, et al. IRE1α: from the function to the potential therapeutic target in atherosclerosis. Mol Cell Biochem. 2024;479(5):1079–1092. doi:10.1007/s11010-023-04780-6
47. Osorio F, Tavernier SJ, Hoffmann E, et al. The unfolded-protein-response sensor IRE-1α regulates the function of CD8α+ dendritic cells. Nat Immunol. 2014;15(3):248–257. doi:10.1038/ni.2808
48. Bergström I, Backteman K, Lundberg A, Ernerudh J, Jonasson L. Persistent accumulation of interferon-γ-producing CD8+CD56+ T cells in blood from patients with coronary artery disease. Atherosclerosis. 2012;224(2):515–520. doi:10.1016/j.atherosclerosis.2012.07.033
49. Hu F, Yu X, Wang H, et al. ER stress and its regulator X-box-binding protein-1 enhance polyIC-induced innate immune response in dendritic cells. Eur J Immunol. 2011;41(4):1086–1097. doi:10.1002/eji.201040831
50. Paulson KE, Zhu SN, Chen M, Nurmohamed S, Jongstra-Bilen J, Cybulsky MI. Resident intimal dendritic cells accumulate lipid and contribute to the initiation of atherosclerosis. Circ Res. 2010;106(2):383–390. doi:10.1161/circresaha.109.210781
51. Xue HM, Sun WT, Chen HX, He GW, Yang Q. Targeting IRE1α-JNK-c-Jun/AP-1-sEH signaling pathway improves myocardial and coronary endothelial function following global myocardial ischemia/reperfusion. Int J Med Sci. 2022;19(9):1460–1472. doi:10.7150/ijms.74533
52. Xiong Y, Yu J. LRRK2 in Parkinson’s disease: upstream regulation and therapeutic targeting. Trends Mol Med. 2024;30(10):982–996. doi:10.1016/j.molmed.2024.07.003
53. Russo I, Bubacco L, Greggio E. LRRK2 as a target for modulating immune system responses. Neurobiol Dis. 2022;169:105724. doi:10.1016/j.nbd.2022.105724
54. Liu Y, Chen L, Gao L, et al. LRRK2 deficiency protects the heart against myocardial infarction injury in mice via the P53/HMGB1 pathway. Free Radic Biol Med. 2022;191:119–127. doi:10.1016/j.freeradbiomed.2022.08.035
55. Mazaki Y, Handa H, Fumoto Y, Horinouchi T, Onodera Y. LRRK2 is involved in the chemotaxis of neutrophils and differentiated HL-60 cells, and the inhibition of LRRK2 kinase activity increases fMLP-induced chemotactic activity. Cell Commun Signal. 2023;21(1):300. doi:10.1186/s12964-023-01305-y
56. Luo J, Thomassen JQ, Nordestgaard BG, Tybjærg-Hansen A, Frikke-Schmidt R. Neutrophil counts and cardiovascular disease. Eur Heart J. 2023;44(47):4953–4964. doi:10.1093/eurheartj/ehad649
57. Manganelli V, Longo A, Mattei V, et al. Role of ERLINs in the Control of Cell Fate through Lipid Rafts. Cells. 2021;10(9):2408. doi:10.3390/cells10092408
58. Manganelli V, Matarrese P, Antonioli M, et al. Raft-like lipid microdomains drive autophagy initiation via AMBRA1-ERLIN1 molecular association within MAMs. Autophagy. 2021;17(9):2528–2548. doi:10.1080/15548627.2020.1834207
59. Li F, Peng J, Lu Y, et al. Blockade of CXCR4 promotes macrophage autophagy through the PI3K/AKT/mTOR pathway to alleviate coronary heart disease. Int J Cardiol. 2023;392:131303. doi:10.1016/j.ijcard.2023.131303
60. Nakagawa K, Tanaka M, Hahm TH, et al. Accumulation of plasma-derived lipids in the lipid core and necrotic core of human atheroma: imaging mass spectrometry and histopathological analyses. Arterioscler Thromb Vasc Biol. 2021;41(11):e498–e511. doi:10.1161/atvbaha.121.316154
61. Xie Z, Fang Z, Pan Z. Ufl1/RCAD, a Ufm1 E3 ligase, has an intricate connection with ER stress. Int J Biol Macromol. 2019;135:760–767. doi:10.1016/j.ijbiomac.2019.05.170
62. Raghunath G, Chen YC, Marin M, Wu H, Melikyan GB. SERINC5-mediated restriction of HIV-1 infectivity correlates with resistance to cholesterol extraction but not with lipid order of viral membrane. Viruses. 2022;14(8):1636. doi:10.3390/v14081636
63. Dybdahl B, Slørdahl SA, Waage A, Kierulf P, Espevik T, Sundan A. Myocardial ischaemia and the inflammatory response: release of heat shock protein 70 after myocardial infarction. Heart. 2005;91(3):299–304. doi:10.1136/hrt.2003.028092
64. Chang YC, Liou JT, Peng YM, Chen GJ, Lin CY, Yang CA. Association of long noncoding RNA expression signatures with stress-induced myocardial perfusion defects. Biomolecules. 2023;13(5):849. doi:10.3390/biom13050849
65. Wong D, Auguste G, Lino Cardenas CL, et al. FHL5 controls vascular disease-associated gene programs in smooth muscle cells. Circ Res. 2023;132(9):1144–1161. doi:10.1161/circresaha.122.321692
66. Grando K, Bessho S, Harrell K, et al. Bacterial amyloid curli activates the host unfolded protein response via IRE1α in the presence of HLA-B27. Gut Microbes. 2024;16(1):2392877. doi:10.1080/19490976.2024.2392877
67. Mączyńska J, Da Pieve C, Burley TA, et al. Immunomodulatory activity of IR700-labelled affibody targeting HER2. Cell Death Dis. 2020;11(10):886. doi:10.1038/s41419-020-03077-6
68. Karayel O, Virreira Winter S, Padmanabhan S, et al. Proteome profiling of cerebrospinal fluid reveals biomarker candidates for Parkinson’s disease. Cell Rep Med. 2022;3(6):100661. doi:10.1016/j.xcrm.2022.100661
69. Bie N, Yong T, Wei Z, et al. Tumor-repopulating cell-derived microparticles elicit cascade amplification of chemotherapy-induced antitumor immunity to boost anti-PD-1 therapy. Signal Transduct Target Ther. 2023;8(1):408. doi:10.1038/s41392-023-01658-3
70. Grabie N, Lichtman AH, Padera R. T cell checkpoint regulators in the heart. Cardiovasc Res. 2019;115(5):869–877. doi:10.1093/cvr/cvz025
71. Liu J, Ma P, Lai L, et al. Transcriptional and immune landscape of cardiac sarcoidosis. Circ Res. 2022;131(8):654–669. doi:10.1161/circresaha.121.320449
72. Sharma S, Plant D, Bowes J, et al. HLA-DRB1 haplotypes predict cardiovascular mortality in inflammatory polyarthritis independent of CRP and anti-CCP status. Arthritis Res Ther. 2022;24(1):90. doi:10.1186/s13075-022-02775-0
73. Mocayar Marón FJ, Camargo AB, Manucha W. Allicin pharmacology: common molecular mechanisms against neuroinflammation and cardiovascular diseases. Life Sci. 2020;249:117513. doi:10.1016/j.lfs.2020.117513
74. Naito AT, Okada S, Minamino T, et al. Promotion of CHIP-mediated p53 degradation protects the heart from ischemic injury. Circ Res. 2010;106(11):1692–1702. doi:10.1161/circresaha.109.214346
75. Qian J, Huo J, Qian J, Gao T, Gou Y. Novel potential therapeutic strategies of senile cardiac amyloidosis: heat shock factor 1 blocks senile cardiac amyloidosis via HSPA1A in mouse model. Int J Cardiol. 2015;195:285–287. doi:10.1016/j.ijcard.2015.05.155
76. Sakabe M, Shiroshita-Takeshita A, Maguy A, et al. Effects of a heat shock protein inducer on the atrial fibrillation substrate caused by acute atrial ischaemia. Cardiovasc Res. 2008;78(1):63–70. doi:10.1093/cvr/cvn019
77. Liu X, Kalogeropulou AF, Domingos S, et al. Discovery of XL01126: a potent, fast, cooperative, selective, orally bioavailable, and blood-brain barrier penetrant PROTAC degrader of leucine-rich repeat kinase 2. J Am Chem Soc. 2022;144(37):16930–16952. doi:10.1021/jacs.2c05499
78. Kapadia P, Bikkina P, Landicho MA, Parekh S, Haas MJ, Mooradian AD. Effect of anti-hyperglycemic drugs on endoplasmic reticulum (ER) stress in human coronary artery endothelial cells. Eur J Pharmacol. 2021;907:174249. doi:10.1016/j.ejphar.2021.174249
79. Wang S, Wang Z, Fan Q, et al. Ginkgolide K protects the heart against endoplasmic reticulum stress injury by activating the inositol-requiring enzyme 1α/X box-binding protein-1 pathway. Br J Pharmacol. 2016;173(15):2402–2418. doi:10.1111/bph.13516
80. Li J, Yue G, Ma W, et al. Ufm1-specific ligase Ufl1 regulates endoplasmic reticulum homeostasis and protects against heart failure. Circ Heart Fail. 2018;11(10):e004917. doi:10.1161/circheartfailure.118.004917
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.