Back to Journals » Journal of Inflammation Research » Volume 18
Identification of Hub Genes for Dexmedetomidine Alleviation of Limb Ischemia-Reperfusion-Induced Lung Injury in Rats by Transcriptomic
Authors Lu K , Ling M, Rao M, Huang H, Liang S, Wei Y , Bai L, Huang Y , Pan L
Received 8 January 2025
Accepted for publication 10 April 2025
Published 24 April 2025 Volume 2025:18 Pages 5427—5445
DOI https://doi.org/10.2147/JIR.S512536
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Qing Lin
Kejian Lu,1– 5,* Maoyao Ling,1,* Mei Rao,6 Haosong Huang,5 Shucong Liang,5 Yanxia Wei,5 Lijuan Bai,5 Yanjuan Huang,5 Linghui Pan1– 4
1Department of Anesthesiology, Guangxi Medical University Cancer Hospital, Nanning, People’s Republic of China; 2Guangxi Engineering Research Center for Tissue and Organ Injury and Repair Medicine, Nanning, People’s Republic of China; 3Guangxi Key Laboratory for Basic Science and Prevention of Perioperative Organ Disfunction, Nanning, People’s Republic of China; 4Guangxi Clinical Research Center for Anesthesiology, Nanning, People’s Republic of China; 5Department of Anesthesiology, The Third Affiliated Hospital of Guangxi Medical University, Nanning, People’s Republic of China; 6Reproductive Medical Center, The Third Affiliated Hospital of Guangxi Medical University, Nanning, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yanjuan Huang, Department of Anesthesiology, The Third Affiliated Hospital of Guangxi Medical University, Nanning, People’s Republic of China, Email [email protected] Linghui Pan, Department of Anesthesiology, Guangxi Medical University Cancer Hospital, Nanning, People’s Republic of China, Email [email protected]
Background: Limb ischemia-reperfusion (LIR), a prevalent clinical condition, frequently precipitates acute lung injury (ALI). Dexmedetomidine (DEX), a selective alpha2-adrenergic receptor agonist, mitigates LIR-induced ALI. However, its underlying mechanisms remain incompletely elucidated. This study aimed to identify hub genes implicated in DEX-mediated protection against LIR-ALI in rats.
Methods: Sprague-Dawley rats were allocated into five groups (n = 3 per group): Sham (femoral artery exposure without occlusion), LIR, LIR + DEX, LIR + Inhibitor, and LIR + DEX + Inhibitor. LIR was induced by clamping the femoral arteries for 3 hours, followed by reperfusion. DEX (50 μg/kg) or Atipamezole (alpha2-receptor inhibitor, 250 μg/kg) was administered prior to ischemia. Lung injury was evaluated via hematoxylin-eosin staining, wet/dry ratio assessment, and quantification of IL-1beta, TNF-alpha, malondialdehyde (MDA), and superoxide dismutase (SOD) levels. RNA sequencing was performed to identify differentially expressed genes (DEGs), followed by functional enrichment analysis, protein-protein interaction (PPI) network construction, and hub gene identification. Gene-gene interaction (GGI) networks were established. Polymerase chain reaction (PCR) and enzyme linked immunosorbent assay (ELISA) validation was conducted.
Results: LIR induced severe lung injury and inflammation, both of which were attenuated by DEX pretreatment. RNA sequencing identified 2,302 DEGs1, 471 DEGs2, 340 DEGs3, and 1,407 DEGs4. After intersection and subtraction analyses, 255 DEX-associated DEGs (DEGs-Dex) and 290 inhibitor-associated DEGs (DEGs-In) were identified, with enrichment in Wnt/PI3K-Akt signaling (DEX) and glycerolipid/butanoate metabolism (In). Nine Hub-Dex genes and four Hub-In genes were identified, among which Selp and Tars1 exhibited a strong positive correlation (correlation = 0.55, P < 0.05). Six hub genes (Tars1, Atf4, Ep300, Sphk1, AABR07051376.1, and Mmp9) were validated.
Conclusion: Six hub genes associated with DEX-mediated protection against LIR-ALI were identified, providing mechanistic insights and potential therapeutic targets for intervention.
Keywords: limb ischemia-reperfusion, acute lung injury, dexmedetomidine, animal model, RNA sequencing, hub genes
Introduction
Limb ischemia-reperfusion (LIR) is a common pathological process in various clinical conditions, including atherosclerotic thrombosis, crush injuries, and surgical interventions, and is associated with significant morbidity and mortality. Ischemia-induced reductions in blood flow result in oxygen and glucose deprivation, leading to metabolic dysfunction and tissue injury. Although reperfusion restores perfusion, it simultaneously triggers severe inflammatory responses, oxidative stress, and immune dysregulation, exacerbating tissue damage, particularly in distant organs such as the lungs. LIR-induced pulmonary injury is characterized by excessive inflammation, oxidative stress, and apoptosis, which, in severe cases, may progress to acute lung injury (ALI) or acute respiratory distress syndrome (ARDS), conditions with mortality rates ranging from 30% to 50%.1
Current therapeutic strategies for LIR include lipid emulsions, Ulinastatin, angiotensin-converting enzyme 2 (ACE2), and p38 Mitogen-activated protein kinases (MAPK) inhibitors, as previously reported. These agents demonstrate potential in attenuating inflammation and oxidative stress; however, their clinical application is hindered by limited efficacy, adverse effects with prolonged use, inadequate prevention of LIR-ALI recurrence, and the need for further clinical validation.2–5 Thus, developing safer and more effective interventions for LIR-ALI remains a pressing clinical challenge.
Dexmedetomidine (DEX), a highly selective alpha2-adrenergic receptor agonist, is widely utilized in anesthesia and intensive care due to its sedative, anxiolytic, analgesic, and hemodynamic-stabilizing properties.6 Recent studies highlight its protective effects against LIR-ALI, demonstrating its ability to modulate inflammation, alleviate oxidative stress, and regulate immune responses.7 DEX suppresses pro-inflammatory cytokine release, enhances alveolar-capillary barrier integrity, and mitigates pulmonary edema and inflammation.8 Moreover, its activation of anti-apoptotic pathways further reduces cellular injury and lung damage.9 Although these findings establish a theoretical framework for DEX-mediated protection in LIR-ALI, its precise molecular mechanisms and clinical applicability require further elucidation through in-depth experimental and clinical investigations.
RNA sequencing (RNA-Seq) has revolutionized transcriptomic analysis by enabling a comprehensive exploration of gene function, regulatory networks, and molecular interactions.10 Unlike traditional gene expression profiling methods, RNA-Seq provides high-resolution genome-wide expression data, facilitating the identification of rare transcripts, alternative splicing variants, and differentially expressed genes with unprecedented accuracy. This approach allows for a holistic investigation of biological processes, offering novel insights into disease pathogenesis and fundamental molecular mechanisms.11 Furthermore, RNA-Seq enhances the detection of gene mutations and regulatory elements, expanding our understanding of dynamic gene expression alterations in response to pathological conditions. Zuo et al12 discovered through RNA-Seq technology that ADAMTS15 may be a potential inflammation-related gene involved in post-conditioning of remote ischemia that confers cardio protection. It may also serve as a potential therapeutic target for future myocardial ischemia-reperfusion injury. This finding provides clues for the treatment of myocardial ischemia-reperfusion injury.
This study established an animal model to assess the protective effects of DEX against LIR-ALI. Using transcriptome sequencing, key biomarkers were identified in lung tissues across experimental groups, and underlying molecular mechanisms were explored. These findings provide valuable insights into LIR-ALI pathophysiology and potential therapeutic targets.
Materials and Methods
Animals
Wild-type male Sprague-Dawley (SD) rats (300 ± 25 g) were obtained from the Animal Experimental Center of Guangxi Medical University. Before experimentation, all rats were acclimated for two weeks in an air-filtered facility under controlled conditions (20–25°C, 50%–70% humidity, 12-hour light/dark cycle) with unrestricted access to food and water. All procedures adhered to the Guiding Opinions on the Treatment of Laboratory Animals and were approved by the Animal Care & Welfare Committee of Guangxi Medical University (#202403016), complying with Chinese government regulations on animal care and use.
Grouping and Limb Ischemia-Reperfusion Rat Model
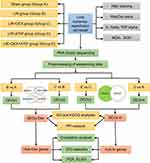
The study design is outlined in Figure 1. Before the experiment, rats were fasted for 4 hours and deprived of water for 1 hour. Anesthesia was induced via intraperitoneal injection of 1% sodium pentobarbital (60 mg/kg), with one-third of the initial dose administered every 45 minutes for maintenance. After induction, the rats were positioned on a heating pad, and an intravenous catheter was inserted into the auricular vein to deliver lactated Ringer’s solution (8–10 mL/h) for hydration.
Rats were randomly assigned to five groups (n = 3 per group): Sham group (group A): Rats received an intraperitoneal injection of 5 mL normal saline. After 30 minutes, the femoral artery was exposed without occlusion. Six hours later, lung and blood samples were collected following euthanasia; LIR group (group B): Ischemia was induced by isolating the femoral artery, applying microvascular clamps, and securing rubber bands at the base of both hindlimbs to block collateral circulation. After 3 hours, the clamps and bands were removed, and reperfusion was allowed. Lung tissue samples were collected 3 hours post-reperfusion; LIR + DEX group (group C): Rats received an intraperitoneal injection of DEX (50 µg/kg, 5 mL, Chia-Tai Tianqing, China, 2309171) 30 minutes before arterial occlusion, followed by the same procedures as group;13,14 LIR + ATIP group (group D): Rats were administered an intraperitoneal injection of atipamezole (alpha2-receptor inhibitor, 250 µg/kg, 5 mL, Macklin, A837199) 30 minutes before arterial occlusion, followed by the same protocol as group;13,14 LIR + DEX + ATIP group (group E): DEX (50 µg/kg) was administered after ATIP (250 µg/kg) immediately, followed by the same steps as group C. Ischemia was confirmed by the absence of dorsal pedal artery pulsation and cold, dark hindlimbs, while reperfusion was verified by restored arterial pulsation, gradual warming, and redness of the hindlimbs.
Histopathological Analysis
The right lower lobe of the lung was fixed in 4% formaldehyde, embedded in paraffin, and sectioned into 4 μm-thick slices, which were mounted on Superfrost Plus microscope slides (Thermo, Waltham, MA). After deparaffinization, sections underwent hematoxylin-eosin (HE) staining for histopathological evaluation. Lung injury severity was assessed based on standard acute lung injury scoring criteria.15 The lung injury scoring system from the American Thoracic Society. It details a scoring system based on five parameters, each with a scale from 0 to 2, indicating the severity of certain lung conditions observed in microscopic fields: A. Neutrophils in the alveolar space: Scored 0 for none, 1 for 1–5 neutrophils, and 2 for more than 5. B. Neutrophils in the interstitial space: Scored 0 for none, 1 for 1–5 neutrophils, and 2 for more than 5. C. Hyaline membranes: Scored 0 for none, 1 for one or more. D. Proteinaceous debris filling the airspaces: Scored 0 for none, 1 for present. E. Alveolar septal thickening: Scored 0 for less than 2 times thickening, 1 for 2 to 4 times, and 2 for more than 4 times. The overall score is calculated using the formula: 
Inflammatory Responses
Pulmonary edema was quantified by determining the wet/dry (W/D) weight ratio. The middle lobe of the right lung was blotted with gauze to remove surface moisture and immediately weighed to obtain the wet weight. The tissue was then dried in a 60°C oven for 48 hours until a constant dry weight was achieved.
Enzyme-Linked Immunosorbent Assays (ELISA)
ELISA is an immunological method based on antigen-antibody binding with high sensitivity, high specificity and easy handling for the identification and quantification of specific proteins in biological samples.16 Inflammatory mediators, including interleukin-1 beta (IL-1beta) and tumor necrosis factor-alpha (TNF-alpha), were measured in lung tissues using commercial ELISA kits (Cusabio, China). Lung homogenates were prepared in 1× phosphate-buffered saline (PBS), and ELISA was performed following the manufacturer’s instructions.
Evaluation of Malondialdehyde (MDA) and Superoxide Dismutase (SOD) in vivo
Oxidative stress markers, including MDA and SOD, were quantified using lipid peroxidation MDA and SOD assay kits (Beyotime, China) according to the manufacturer’s protocols.
RNA Library Construction and Sequencing
Total RNA was extracted following the standard RNA-Seq library construction protocol. Trizol reagent (ThermoFisher, USA, 15596018) was used for RNA isolation, and sample quality and integrity were assessed using the Bioanalyzer 2100 and RNA 6000 Nano LabChip kit (Agilent, USA, 5067–1511). Samples with RNA integrity number (RIN) values ≤7.0 were excluded. mRNA was purified through two rounds of selection, followed by fragmentation using the Magnesium RNA Fragmentation Module (NEB, USA, cat.e6150) at 94°C for 5–7 minutes. First- and second-strand cDNA synthesis was performed using SuperScript™ II reverse transcriptase (Invitrogen, USA, cat.1896649), E. coli DNA polymerase I (NEB, USA, cat.m0209), RNase H (NEB, USA, cat.m0297), and dUTP solution (ThermoFisher, USA, cat.R0133). After end repair and adapter ligation, PCR amplification was performed to generate cDNA libraries with an average fragment size of 300 ± 50 bp. Paired-end sequencing (PE150) was conducted using the Illumina NovaSeq™ 6000 platform.
Preprocessing of Sequencing Data
Raw sequencing reads were processed using fastp to remove adaptor sequences, low-quality bases, and undetermined nucleotides. Cleaned reads were mapped to the Rattus norvegicus reference genome (Ensembl_v104) using HISAT2. Transcript assembly was performed using StringTie with default parameters, followed by transcriptome reconstruction with gffcompare. Expression levels of all transcripts were quantified using StringTie, with fragments per kilobase of transcript per million mapped reads (FPKM) calculated to assess mRNA expression.
Principal Component Analysis (PCA)
Based on sequencing data, PCA and orthogonal partial least squares discriminant analysis (OPLS-DA) were performed on 15 samples from different groups using the MetaboAnalyst package (version 2.0.0).17
Identification of Differentially Expressed Genes (DEGs)
To analyse the mitigating effect of DEX on LIR- ALI in rats, the data were standardised using the median ratio method of the DESeq2 software package (version 1.38.0) and analysed for differences.18 First, comparisons were made between the Sham group (group A) and the LIR group (group B) (B vs A), as well as between the LIR group (group B) and the LIR + DEX group (group C) (C vs B), with thresholds set at |log2 fold change (FC)| > 0.5 and P < 0.05. The resulting DEGs were designated as DEGs1 and DEGs2, respectively. Using the VennDiagram package (version 1.7.3),19 DEGs associated with DEX-mediated protection were identified by intersecting the upregulated DEGs1 with the downregulated DEGs2, and the downregulated DEGs1 with the upregulated DEGs2. These genes, representing the transcriptional response to DEX treatment, were labeled as DEGs-Dex.
To further examine the inhibitory effect of ATIP on DEX-mediated lung protection in LIR rats, additional DEG analyses were performed using DESeq2. Comparisons were made between the LIR + DEX group (group C) and the LIR + DEX + ATIP group (group E) (E vs C), as well as between the LIR group (group B) and the LIR + ATIP group (group D) (D vs B), with the same cutoff criteria (|log2FC| > 0.5, P < 0.05). The resulting DEGs were designated as DEGs3 and DEGs4, respectively. To identify genes involved in ATIP-mediated reversal of DEX protection, DEGs4 was subtracted from DEGs3, retaining only the remaining genes of DEGs3. These genes were labeled as DEGs-In.
For visualization, volcano plots were generated for all DEG analyses using the ggplot2 package (version 3.4.4),20 highlighting the top 10 upregulated and downregulated genes based on |log2FC|. Additionally, heatmaps of DEGs across different groups were constructed using the ComplexHeatmap package (version 2.14.0).21
Enrichment Analysis
To elucidate the functional roles and signaling pathways associated with DEGs-Dex and DEGs-In, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed using the clusterProfiler package (version 4.7.1.3),22 with significance set at P < 0.05. GO and KEGG results were ranked in ascending order of P values, and the top 10 enriched terms were visualized. KEGG pathway results were further depicted using GO plot (version 1.0.2),23 which generated chord diagrams to illustrate key functional associations. In addition, the ReactomePA (version 4.7.1.3)24 was employed to re-explore the pathways of Hub-Dex genes and Hub-In genes based on the Reactome database (P < 0.05).
Construction of Protein-Protein Interaction (PPI) Network
Subsequently, DEGs-Dex and DEGs-In were separately input into STRING (confidence score = 0.4) to analyze PPI. The resulting PPI networks were then visualized using Cytoscape (version 3.7.2).25
Acquisition of Hub Genes
To identify hub genes, the CytoHubba plugin in Cytoscape (version 3.7.1) was used to analyze DEGs-Dex with five ranking algorithms: Betweenness, MCC, Degree, EPC, and MNC. The top 20 genes from each algorithm were selected, and their intersections were determined to identify hub genes (Hub-Dex genes and Hub-In genes). UpSetR (version 1.4.0) was employed to generate upset plots visualizing these intersections.26
Correlation Analysis
Spearman correlation analysis was conducted using the psych package (version 2.4.3), with a threshold of |correlation (cor)| > 0.3 and P < 0.05, to explore relationships between Hub-Dex and Hub-In genes.
Construction of Gene-Gene Interaction Networks (GGIs)
GGI networks were constructed separately for Hub-Dex and Hub-In genes using the GeneMANIA algorithm to elucidate their functional associations.
Verification of Hub Genes
Total RNA was extracted from the left lung tissue (n = 6 per group) using TRIzol reagent (Invitrogen, USA). Tissue samples were homogenized, incubated on ice for 5 minutes, and subjected to phase separation with chloroform, followed by vortexing and centrifugation. The aqueous phase was transferred to a clean Eppendorf (EP) tube, precipitated with isopropanol, and centrifuged. After discarding the supernatant, the RNA pellet was washed with 75% ethanol, centrifuged, air-dried, and resuspended in RNase-free water. The dissolved RNA was incubated at 4°C for 10 minutes, and its concentration was measured using a NanoDrop spectrophotometer (Thermo Fisher, USA). cDNA synthesis was performed using a reverse transcription kit (Takara, Japan) following the manufacturer’s protocol. The PCR reaction mix was prepared with TB Green Premix Ex Taq II (Takara, Japan) and processed on a qTOWER real-time PCR system (Analytik Jena, Germany). Relative gene expression was quantified using the 2−ΔΔCT method, with beta-actin as an internal reference. Primers were synthesized by Sangon Biotech (Shanghai, China), and their sequences are listed in Table 1. The PCR cycling conditions included an initial denaturation at 95°C for 30 seconds, followed by 40 cycles of 95°C for 5 seconds and 60°C for 10 seconds for annealing/extension.
 |
Table 1 Primer Sequences for PCR |
ELISA kits (UpingBio, China) were measured the protein levels of Hub genes in serum and lung tissues (n = 6 per group). Lung homogenates were prepared using 1 × PBS, and the ELISA was performed according to the manufacturer’s instructions.
Statistical Analysis
Bioinformatics analyses were conducted using R (version 4.2.2). Data processing and statistical analyses were performed using SPSS (version 22.0, SPSS Inc., USA). One-way analysis of variance (ANOVA) was applied for comparisons, with P < 0.05 considered statistically significant. The post-hoc test was conducted using Honestly Significant Difference (Tukey’s HSD) method.
Results
Animal-Related Experiments
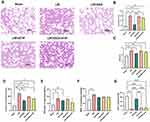
Compared to the Sham group, the LIR group exhibited severe lung injury, characterized by alveolar septal thickening, pulmonary edema, alveolar structural disruption, and inflammatory cell infiltration, as revealed by HE staining. DEX pretreatment significantly alleviated these pathological alterations, whereas lung injury in the LIR + ATIP and LIR + DEX + ATIP groups remained comparable to that observed in the LIR group (Figure 2A). Markers of lung injury and inflammation were significantly elevated in the LIR group relative to the Sham group, including increased lung injury scores (Figure 2B), W/D ratios (Figure 2C), elevated levels of IL-1beta, TNF-alpha, and MDA, and reduced SOD activity (Figure 2D–G). These results indicate that LIR induces substantial pulmonary pathology and inflammation, contributing to LIR-ALI, while DEX pretreatment effectively mitigates these lung injuries.
PCA
Analysis of sequencing data from five experimental groups revealed significant intergroup differences, confirming the high quality of sequencing data and justifying further investigation (Figure 3).
Identification of DEGs-Dex and DEGs-In
Differential expression analysis identified 2,302 DEGs1 in the LIR group (upregulated: 1,376; downregulated: 926), 471 DEGs2 in the LIR + DEX group (upregulated: 195; downregulated: 276), 340 DEGs3 in the LIR + DEX + ATIP group (upregulated: 162; downregulated: 178), and 1,407 DEGs4 in the LIR + ATIP group (upregulated: 527; downregulated: 880) (P < 0.05) (Figure 4A–D) Heatmaps generated for DEGs1, DEGs2, DEGs3, and DEGs4 demonstrated significant expression differences between groups, enabling clear distinction between experimental conditions (Figure 4E–H). To identify DEX-responsive genes, upregulated DEGs1 were intersected with downregulated DEGs2, while downregulated DEGs1 were intersected with upregulated DEGs2, yielding 255 DEGs-Dex (Figure 5A and B). Similarly, DEGs3 and DEGs4 were subtracted to identify 290 DEGs-In, representing genes involved in ATIP-mediated reversal of DEX’s protective effects (Figure 5C).
Function Enrichment of DEGs-Dex and DEGs-In
GO analysis identified 331 biological processes (BP) (P < 0.05) associated with DEGs-Dex, including regulation of cell growth, Wnt signaling pathway, and cell-cell signaling by Wnt (Figure 6A and Supplementary Table 1). KEGG enrichment analysis revealed 210 pathways related to DEGs-Dex, with key pathways including sphingolipid metabolism, PI3K-Akt signaling pathway and Wnt signaling (Figure 6B and Supplementary Table 2). For DEGs-In, 13 GO terms were identified, comprising one cellular component (CC) and 12 molecular functions (MF), such as glycoprotein complex and cytokine binding (Figure 6C and Supplementary Table 3). Additionally, 185 KEGG pathways were associated with DEGs-In, including glycerolipid metabolism, cytokine-cytokine receptor interaction, and butanoate metabolism (Figure 6D and Supplementary Table 4). Finally, using the Reactome database, it was found that DEGs-Dex were enriched for 54 pathways, including RIP-mediated NF-kappa B activation via ZBP1, Sphingolipid metabolism (Supplementary Figure 1A and Supplementary Table 5), while DEGs-In were enriched for 14 pathways, such as Interleukin-12 family signaling, EPH-ephrin mediated repulsion of cells (Supplementary Figure 1B and Supplementary Table 6).
Acquisition of Hub-Dex Genes and Hub-In Genes
The PPI network constructed using DEGs-Dex comprised 107 nodes and 100 edges, featuring key genes such as Ep300 (Figure 7A). In contrast, the PPI network derived from DEGs-In contained 100 nodes and 125 edges, including Mmp9 (Figure 7B). Intersection analysis of the top 20 genes identified by five ranking algorithms resulted in nine Hub-Dex genes (Tars1, Atf4, Rps25, Ep300, Sphk1, Itga4, Itgb8, Farsb, and AABR07051376.1) and four Hub-In genes (Mmp9, Selp, Cxcl6, and Ace3) (Figure 7C and D).
Correlation Analysis of Hub Genes and GGI Networks Construction
Correlation analysis between Hub-Dex and Hub-In genes revealed a significant positive correlation between Selp and Tars1 (cor = 0.55, P < 0.05) (Figure 8A) (Supplementary Table 7). Further examination of the GGI network constructed from Hub-Dex genes and their neighboring genes demonstrated that these hub genes were functionally associated through shared protein domains and were involved in sphingolipid metabolism, membrane lipid metabolism, and tRNA aminoacylation for protein translation (Figure 8B). Meanwhile, the GGI network of Hub-In genes primarily exhibited co-expression patterns, with enrichment in biological processes such as the cellular response to biotic stimuli and response to lipopolysaccharides (Figure 8C).
PCR Verification Result
mRNA expression analysis indicated that Tars1, Atf4, Ep300, Sphk1, and Mmp9 were significantly upregulated in the LIR group compared to the Sham group (P < 0.05). Notably, DEX treatment markedly reduced the expression levels of Tars1, Atf4, Sphk1, AABR07051376.1, and Mmp9 relative to the LIR group (P < 0.05) (Figure 9).
ELISA Verification Result
Due to the lack of specific ELISA kits for Tars1 and AABR07051376.1, we focused on measuring the expression levels of Atf4, Ep300, Sphk1, and Mmp9 in serum and lung tissue of rats. ELISA results demonstrated that the levels of these four genes were significantly elevated in both serum and lung tissue in the LIR group compared to the Sham group, whereas DEX preconditioning markedly suppressed their expression (Figure 10).
Discussion
LIR injury is a complex pathological condition driven by intricate molecular and cellular mechanisms that lead to systemic organ damage, including lung injury. The protective effects of DEX, especially its antioxidant and anti-inflammatory properties in ameliorating IR injury, have been demonstrated in previous studies.27,28 This study aimed to elucidate the molecular processes underlying LIR-ALI through a comprehensive experimental approach combining animal modeling and multi-omics analysis. By utilizing sequencing data from five rat groups—Sham, LIR, LIR + DEX, LIR + inhibitor, and LIR + DEX + inhibitor—differential gene expression, functional enrichment, PPI networks, hub gene identification, and subsequent validation were explored via PCR and ELISA. This integrated analysis offers novel insights into the mechanisms by which DEX mitigates LIR-ALI and identifies potential therapeutic targets.
The rat LIR model effectively recapitulates LIR-ALI, as confirmed by histopathological examination, lung injury scores, and markers of inflammation and oxidative stress. In LIR injury, free radicals induce lipid peroxidation of cell membranes, with MDA serving as a terminal product and a biomarker of LIR-induced damage.29 SOD, a key antioxidant enzyme, scavenges free radicals and exerts a protective effect against ischemia-reperfusion injury;30 however, its activity is significantly reduced in LIR injury models.31,32 In this study, compared to the Sham group, rats in the LIR model group exhibited increased levels of IL-1beta, TNF-alpha, and MDA, along with decreased SOD activity. Histopathological examination further revealed alveolar septal thickening, pulmonary edema, and disruption of alveolar structures, confirming the successful establishment of the LIR-ALI rat model. This model is highly reliable and reproducible, making it a valuable tool for investigating the pathophysiological changes and therapeutic mechanisms associated with LIR-induced lung damage.14 Furthermore, in the animal model experiments, we observed that the levels of IL-1beta and the lung injury score in the LIR+DEX group, were significantly reduced compared to the LIR group, indicating that DEX has a protective effect against LIR. Further research found that the SOD activity and lung injury score in the LIR + DEX + ATIP group were not significantly different from those in the LIR group and the LIR + ATIP group, suggesting that the protective effect of DEX on the lungs may be partially mediated by the alpha2-adrenergic receptor. It is worth noting that the level of IL-1beta in the LIR + DEX + ATIP group increased compared to the LIR + DEX group but was still significantly lower than in the LIR group, indicating that the inhibitory effect of DEX on IL-1beta may not be entirely dependent on the alpha2-adrenergic receptor. We speculate that the therapeutic effect of DEX on LIR may involve multiple mechanisms, some of which are independent of the alpha2-adrenergic receptor signaling pathway. More research is still needed in the future to further explore and clarify its specific molecular mechanisms.
In this experiment, rats were divided into Sham, LIR, and various drug intervention groups. By implementing a comprehensive treatment protocol, the study assessed the impact of different drugs on LIR-ALI. The multi-group comparison design enables a detailed analysis of the mechanisms underlying the effects of individual drugs and their combinations, particularly focusing on the lung-protective roles of DEX and inhibitors such as Atipamezole. This study advances our understanding of the mechanisms through which DEX confers lung protection, offering potential clinical implications for preventing and treating LIR-ALI.
Functional enrichment analysis revealed key biological processes, such as regulation of vasculature development and cell growth regulation, which are essential for the recovery of lung and other organ functions following IR injury. Previous research has identified microvascular dysfunction as a pivotal factor in LIR injury, with repair processes reliant on cytokine regulation and their effects on cellular states.33,34 These findings further corroborate the validity of the enrichment analysis.
KEGG analysis revealed that DEGs-Dex modulate 210 signaling pathways, including sphingolipid metabolism, the Wnt signaling pathway and PI3K-Akt signaling pathway. These pathways not only play pivotal roles in inflammation regulation but are also closely associated with hemostasis and tissue repair.Sphingosine-1-phosphate (S1P), a key molecule in sphingolipid metabolism, has been reported to protect against IR injury by regulating vascular permeability and inflammatory cell migration.35 Moreover, S1P is implicated in both hemostasis and tissue repair through its dual role in promoting endothelial barrier function and inhibiting inflammatory cell infiltration.36,37 This pathway provides valuable insights into the repair mechanisms of lung injury induced by IR. Similarly, the Wnt signaling pathway has been shown to be reactivated in cardiac IR injury28 and is considered a critical therapeutic target in cerebral IR injury.38
The PI3K-Akt signaling pathway is a key regulator of cellular apoptosis and inflammatory responses.39,40 DEX has been shown to significantly enhance PI3K/Akt protein activation in response to LPS stimulation41 and effectively mitigate ischemia/reperfusion-induced lung injury via activation of the PI3K/Akt/HIF-1alpha signaling axis.42 Consistently, our findings further support the protective role of DEX in IR-induced lung injury. Furthermore, our study identified a significant downregulation of Atf4 expression in the DEX-treated group compared to the model group. As a key downstream node of the PI3K/Akt/mTOR signaling cascade, Atf4 exhibits anti-inflammatory potential.43 Knockdown of Atf4 has been shown to markedly reduce pro-inflammatory cytokines IL-1beta, IL-6, and TNF-alpha, while concomitantly increasing the expression of the anti-inflammatory cytokine IL-10.44 These findings suggest that DEX may exert its anti-inflammatory and protective effects through the regulation of the PI3K/Akt pathway and its downstream effector Atf4. However, the precise mechanisms underlying DEX-mediated modulation of Atf4 expression and its interactions with other signaling networks warrant further investigation. Collectively, sphingolipid metabolism, the Wnt signaling pathway, and the PI3K-Akt signaling pathway are intricately linked to the pathophysiology of IR injury. DEX may confer protection by orchestrating the dynamic balance between hemostasis, tissue repair, and inflammation suppression. Nevertheless, further studies are required to elucidate the precise regulatory mechanisms of DEX on these pathways and their interplay under different pathological conditions, ultimately providing a more comprehensive theoretical framework for IR injury therapy.
Additionally, GO enrichment analysis identified functions related to glycoprotein complexes and cytokine binding, suggesting that these molecules may contribute to regulating inflammation and promoting tissue repair by mediating cell adhesion and inflammatory signaling.45,46 These results emphasize the critical importance of inflammation regulation, metabolic homeostasis, and signal transduction in the context of LIR-induced lung injury.
Functional enrichment analysis of DEGs-In further revealed significant enrichment in 185 signaling pathways, including glycerolipid metabolism, cytokine-cytokine receptor interactions, and butanoate metabolism. Glycerolipid metabolism has been implicated in influencing inflammation and apoptosis by modulating intracellular lipid signaling. During IR injury, disruptions in phospholipid metabolism in type II alveolar epithelial cells may impair surfactant function, exacerbating lung injury.47 Furthermore, chemokines such as Monocyte chemoattractant protein-1 (MCP-1) and Macrophage inflammatory protein-2 (MIP-2), involved in cytokine-cytokine receptor interactions, increase vascular permeability and promote edema formation.48 Comparison of pathways significantly enriched in DEGs-In and DEGs-Dex revealed a shared enrichment in the cytokine–cytokine receptor interaction pathway, suggesting that this pathway may be regulated by both DEX and the alpha2-adrenergic receptor inhibitor (ATIP). Butyrate, a short-chain fatty acid (SCFA), regulates immune cell activity through G protein-coupled receptors (GPRs) like GPR109A, reducing neutrophil infiltration and mitigating the severity of lung injury.49
PPI network analysis identified key nodes in the DEGs-Dex and DEGs-In networks. The DEGs-Dex network pinpointed E1A binding protein p300 (Ep300) as a central hub, while the DEGs-In network highlighted Mmp9 as a critical gene. Ep300, an essential epigenetic regulator, mediates histone acetylation and gene transcription via chromatin remodeling. Acetylation by Ep300 has been implicated in the activation of the NOD-like receptor protein 3 (NLRP3) inflammasome. This process exacerbates inflammation and tissue damage during IR injury by promoting the transcription of inflammasome components and downstream cytokines, such as IL-1beta and IL-18.50 Similarly, matrix metalloprotein 9 (Mmp9), identified as a key gene in the DEGs-In network, is crucial in inflammation and tissue repair. Overexpression of Mmp9, induced by hypoxia and oxidative stress during IR, contributes to increased capillary permeability through degradation of the basement membrane and endothelial tight junctions, leading to tissue edema and inflammatory cell infiltration, particularly in lung injury models.51 These hub genes likely interact to facilitate tissue repair following IR injury, offering potential molecular targets for therapeutic intervention.
Correlation analysis revealed significant positive correlations between Hub-Dex and Hub-In genes. For example, the correlation between Selectin P (Selp) and Threonyl-TRNA Synthetase 1 (Tars1) reached 0.55, with a P < 0.05. Selp, an adhesion molecule primarily expressed on activated platelets and endothelial cells, plays a pivotal role in leukocyte adhesion and migration during inflammation. Tars1, involved in tRNA aminoacylation, has not been previously associated with Selp. This correlation suggests a potential co-regulation through nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kappa B) or MAPK signaling pathways, which could further drive the release of pro-inflammatory cytokines.
In the constructed GGI network, analysis of the Hub-Dex genes and their neighboring genes revealed co-expression of multiple shared protein domains, with involvement in biological processes such as sphingolipid metabolism, membrane lipid metabolism, and tRNA aminoacylation during RNA translation. These results indicate functional overlap between Hub-Dex genes and adjacent genes in metabolic and translational pathways induced by LIR, providing novel insights into the molecular mechanisms underlying LIR injury. In the GGI network of Hub-In genes, the co-expressed genes were predominantly associated with cellular responses to biological stimuli and lipopolysaccharide (LPS) responses, highlighting the close connection of Hub-In genes with inflammation and immune responses during LIR. The network constructed with GeneMANIA revealed the intricate interactions between these Hub genes, emphasizing their collaborative roles in inflammation, cell migration, and stress responses.
Atf4, a Hub-Dex gene, has attracted considerable attention for its role in IR injury. As a key mediator of endoplasmic reticulum (ER) stress responses, Atf4 regulates cellular responses to oxidative and ER stress, crucial for maintaining cell homeostasis and survival under ischemic conditions.52,53 IR injury activates both ER and oxidative stress, with Atf4 modulating endothelial tight junction proteins (eg, claudin-5 and occludin), influencing blood-brain barrier (BBB) and microvascular endothelial permeability.54 For instance, Atf4 inhibition has been shown to alleviate renal IR injury,55 and CCAAT/enhancer binding protein alpha (C/EBP alpha)-mediated deacetylation of Atf4 through Histone Deacetylase 1 (HDAC1) has been reported to suppress ER stress, reducing hepatic IR injury.56 The findings corroborate our results, where Atf4 is upregulated in the LIR group compared to the Sham group, and downregulated in the LIR + Dex group, indicating that Dex exerts a protective effect against LIR.
Similarly, Sphk1 has been implicated in IR injury. By catalyzing the conversion of sphingosine to S1P, Sphk1 is integral to angiogenesis and immune cell function. In LIR-ALI, Sphk1 regulates the S1P signaling pathway, modulating vascular permeability, cell migration, and inflammation, thus facilitating lung injury repair.57 The Sphk1/S1P axis may exert protective effects in IR-ALI through enhanced angiogenesis and immune response modulation,58 positioning Sphk1 as a potential therapeutic target for LIR-ALI. Studies have suggested that the Sphk1/S1P pathway may confer protection against IR-induced acute lung injury (IR-ALI) by enhancing angiogenesis and modulating immune responses.59 Moreover, Sphk1 serves as a negative regulator of platelet function, inhibiting platelet degranulation, aggregation, and thrombosis, which may help alleviate microvascular dysfunction in IR injury.59 Further animal studies have demonstrated that Sphk1-knockout mice exhibit delayed wound healing, impaired angiogenesis, and defective inflammatory cell recruitment.60 These findings underscore the therapeutic potential of Sphk1 in LIR-ALI, suggesting that it may promote tissue repair and resolution of inflammation through multifaceted mechanisms. Targeting the Sphk1/S1P pathway could thus offer a novel strategy for the treatment of IR-ALI.
Ep300 is crucial in LIR-ALI by modulating various biological processes. It regulates transcription factors such as NF-kappa B and activator protein-1 (AP-1), central mediators of the inflammatory response.61 Ep300, through acetylation, amplifies the expression of pro-inflammatory cytokines like TNF-alpha and IL-1beta, key drivers of IR injury pathophysiology. Notably, the interaction between Ep300 and NF-kappa B has been linked to heightened inflammation under conditions of hypoxia and oxidative stress following IR injury.
Mmp9, a matrix metalloproteinase, is involved in extracellular matrix remodeling and promotes inflammatory cell recruitment, contributing to pro-inflammatory responses and tissue damage in LIR injury.62 Recent studies have underscored Mmp9’s role in IR injury, particularly in lung tissue damage, where it facilitates neutrophil migration and inflammatory processes, leading to extracellular matrix remodeling and tissue degradation.63 Upregulation of Mmp9 expression correlates with increased alveolar barrier dysfunction and vascular permeability, exacerbating post-ischemic damage.64 These findings align with PCR data demonstrating Mmp9 overexpression in the LIR-ALI model, highlighting its critical role in the inflammatory response and tissue destruction following ischemic injury.
Tars1 and AABR07051376.1 remain underexplored in the context of LIR injury, with their roles still being investigated. Emerging evidence indicates that Tars1 may modulate inflammatory responses through interactions with critical signaling pathways, such as NF-kappa B, suggesting its involvement in regulating inflammation and apoptosis following ischemic injury.65 AABR07051376.1, also known as Ppp1r26, is a long non-coding RNA (lncRNA) that has recently gained attention due to its association with tumor malignancy and prognosis in cancers such as breast and liver cancer.66 Further research is needed to clarify their contributions to LIR pathophysiology.
In this study, we employed genomic methods to identify hub genes involved in the rat LIR lung injury model. However, there are some limitations. First, the sample size used for RNA sequencing was small, which may affect the stability of the results. Second, although we validated the expression changes of hub genes through PCR and ELISA, we lack direct functional validation and studies on regulatory mechanisms. Moreover, the current results are limited to the rat model and have not been verified in clinical samples. Therefore, we plan to expand the sample size in future studies and use CRISPR/Cas9 gene-editing technology to construct gene knockout models. We will also combine single-cell sequencing with multi-omics integrated analysis to systematically and deeply explore the protective mechanisms of DEX. Additionally, we will collect clinical samples to assess the translational value of our findings, providing a more reliable theoretical basis for the clinical treatment of LIR-ALI.
Conclusion
In conclusion, a lung injury model induced by LIR was established, and transcriptomic analysis revealed biomarkers linked to DEX-mediated alleviation of LIR-ALI. After experimental validation, six potential hub genes (Tars1, Atf4, Ep300, Sphk1, Aabr07051376.1, and Mmp9) were identified, providing new insights into therapeutic targets for DEX in LIR-ALI.
Data Sharing Statement
The original contributions presented in the study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding authors.
Ethical Approval
The current study was performed with the approval of the Ethics Committee of The Animal Care & Welfare Committee of Guangxi Medical University. The animal experiment was conducted in accordance with Guiding Opinions on the Treatment of Laboratory Animals issued by the Ministry of Science and Technology of the People’s Republic of China.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by Guangxi Clinical Research Center for Anesthesiology (#GK AD22035214) and Middle-aged and Young Teachers’ Basic Ability Promotion Project of Guangxi (#2024KY0131).
Disclosure
The authors declare that there are no conflicts of interest regarding the publication of this paper.
References
1. Bellani G, Laffey JG, Pham T, et al. Epidemiology, patterns of care, and mortality for patients with acute respiratory distress syndrome in intensive care units in countries. JAMA. 2016;315(8):788–800. doi:10.1001/jama.2016.0291
2. Xia FF, Xia Y, Chen SS, et al. Lipid emulsion mitigates impaired pulmonary function induced by limb ischemia/reperfusion in rats through attenuation of local cellular injury and the subsequent systemic inflammatory. BMC Anesthesiol. 2017;17(1):121. doi:10.1186/s12871-017-0375-6
3. Wu J, Yan X, Jin GQ. Ulinastatin protects rats from sepsis-induced acute lung injury by suppressing the JAK-STAT3 pathway. J Cell Biochem. 2019;120(2):2554–2559. doi:10.1002/jcb.27550
4. Ye RS, Liu ZW. ACE2 exhibits protective effects against LPS-induced acute lung injury in mice by inhibiting the LPS-TLR4 pathway. Exp Mol Pathol. 2020;113. doi:10.1016/j.yexmp.2019.104350
5. Wang T, Liu C, Pan LH, et al. Inhibition of p38 MAPK mitigates lung ischemia reperfusion injury by reducing blood-air barrier hyperpermeability. Front Pharmacol. 2020;11:569251. doi:10.3389/fphar.2020.569251
6. Wang K, Wu M, Xu J, et al. Effects of dexmedetomidine on perioperative stress, inflammation, and immune function: systematic review and meta-analysis. Br J Anaesth. 2019;123(6):777–794. doi:10.1016/j.bja.2019.07.027
7. Ciftel S, Mercantepe F, Mercantepe T, et al. Dexmedetomidine on the interplay of IL-6 and STAT3 pathways in adrenal gland damage-induced scalding burns in rats. Naunyn Schmiedebergs Arch Pharmacol. 2024;397:15. doi:10.1007/s00210-024-03300-7
8. Liu J, Huang XH, Hu SP, et al. Dexmedetomidine attenuates lipopolysaccharide-induced acute lung injury in rats by inhibition of caveolin-1 downstream signaling. Biomed Pharmacother. 2019;110:118. doi:10.1016/j.biopha.2019.109314
9. Shi J, Yu TX, Song K, et al. Dexmedetomidine ameliorates endotoxin-induced acute lung injury in vivo and in vitro by preserving mitochondrial dynamic equilibrium through the HIF-1a/HO-1 signaling pathway. Redox Biol. 2021:41. doi:10.1016/j.redox.2021.101954
10. Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi:10.1038/nrg2484
11. Li T, Lin X, Yu L, et al. RNA-seq profiling of Fugacium kawagutii reveals strong responses in metabolic processes and symbiosis potential to deficiencies of iron and other trace metals. Sci Total Environ. 2020;705:135767. doi:10.1016/j.scitotenv.2019.135767
12. Zuo B, Zhu S, Wang G, Li Z. Transcriptome analysis reveals ADAMTS15 is a potential inflammation-related gene in remote ischemic postconditioning. Front Cardiovasc Med. 2023;10:1089151. doi:10.3389/fcvm.2023.1089151
13. Xue BB, Chen BH, Tang YN, et al. Dexmedetomidine protects against lung injury induced by limb ischemia-reperfusion via the TLR4/MyD88/NF-κB pathway. Kaohsiung J Med Sci. 2019;35(11):672–678. doi:10.1002/kjm2.12115
14. Wang L, Ding Y, Bai Y, et al. The activation of SIRT3 by dexmedetomidine mitigates limb ischemia-reperfusion-induced lung injury. Ann Transl Med. 2022;10(6):319. doi:10.21037/atm-22-711
15. Matute-Bello G, Downey G, Moore BB, et al. An official American Thoracic Society workshop report: features and measurements of experimental acute lung injury in animals. Am J Respir Cell mol Biol. 2011;44(5):725–738. doi:10.1165/rcmb.2009-0210ST
16. Lenz KD, Klosterman KE, Mukundan H, et al. Lipoprotein capture ELISA method for the sensitive detection of amphiphilic biomarkers. Anal Biochem. 2022;652:114747. doi:10.1016/j.ab.2022.114747
17. Chong J, Xia J. MetaboAnalystR: an R package for flexible and reproducible analysis of metabolomics data. Bioinformatics. 2018;34(24):4313–4314. doi:10.1093/bioinformatics/bty528
18. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi:10.1186/s13059-014-0550-8
19. Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 2011;12:35. doi:10.1186/1471-2105-12-35
20. Gustavsson EK, Zhang D, Reynolds RH, et al. ggtranscript: an R package for the visualization and interpretation of transcript isoforms using ggplot2. Bioinformatics. 2022;38(15):3844–3846. doi:10.1093/bioinformatics/btac409
21. Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–2849. doi:10.1093/bioinformatics/btw313
22. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287. doi:10.1089/omi.2011.0118
23. Walter W, Sánchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31(17):2912–2914. doi:10.1093/bioinformatics/btv300
24. Yu G, He QY. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst. 2016;12(2):477–479. doi:10.1039/c5mb00663e
25. Otasek D, Morris JH, Bouças J, et al. Cytoscape automation: empowering workflow-based network analysis. Genome Biol. 2019;20(1):185. doi:10.1186/s13059-019-1758-4
26. Conway JR, Lex A, Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33(18):2938–2940. doi:10.1093/bioinformatics/btx364
27. She H, Hu Y, Zhao GZ, et al. Dexmedetomidine ameliorates myocardial ischemia-reperfusion injury by inhibiting MDH2 lactylation via regulating metabolic reprogramming. Adv Sci. 2024;11. doi:10.1002/advs.202409499
28. Yang YF, Wang H, Song N, et al. Dexmedetomidine attenuates ischemia/reperfusion-induced myocardial inflammation and apoptosis through inhibiting endoplasmic reticulum stress signaling. J Inflamm Res. 2021;14:1217–1233. doi:10.2147/jir.S292263
29. Ince İ, Akar İ, Arıcı A. Renoprotective effect of edaravone in acute limb ischemia/reperfusion injury. Turk Gogus Kalp Damar Cerrahisi Derg. 2019;28(2):274–281. doi:10.5606/tgkdc.dergisi.2020.18905
30. Li R, Fan L, Ma F, et al. Effect of etomidate on the oxidative stress response and levels of inflammatory factors from ischemia-reperfusion injury after tibial fracture surgery. Exp Ther Med. 2017;13(3):971–975. doi:10.3892/etm.2017.4037
31. Nazli Y, Colak N, Namuslu M, et al. Cilostazol attenuates spinal cord ischemia-reperfusion injury in rabbits. J Cardiothorac Vasc Anesth. 2015;29(2):351–359. doi:10.1053/j.jvca.2014.06.028
32. Zhang Y, Xu H, Wang T, et al. Remote limb ischemic post-conditioning attenuates ischemia-reperfusion injury in rat skin flap by limiting oxidative stress. Acta Cir Bras. 2016;31(1):15–21. doi:10.1590/s0102-865020160010000003
33. Chen F, Zhan J, Liu M, et al. FGF2 alleviates microvascular ischemia-reperfusion injury by KLF2-mediated ferroptosis inhibition and antioxidant responses. Int J Biol Sci. 2023;19(13):4340–4359. doi:10.7150/ijbs.85692
34. Chou HC, Chan HL. 5-Methoxytryptophan-dependent protection of cardiomyocytes from heart ischemia reperfusion injury. Arch Biochem Biophys. 2014;543:15–22. doi:10.1016/j.abb.2013.12.014
35. Borodzicz-Jażdżyk S, Jażdżyk P, Łysik W, et al. Sphingolipid metabolism and signaling in cardiovascular diseases. Front Cardiovasc Med. 2022;9:915961. doi:10.3389/fcvm.2022.915961
36. Mahajan-Thakur S, Böhm A, Jedlitschky G, et al. Sphingosine-1-phosphate and its receptors: a mutual link between blood coagulation and inflammation. Mediators Inflamm. 2015;2015:831059. doi:10.1155/2015/831059
37. Chen T, Song P, He M, et al. Sphingosine-1-phosphate derived from PRP-Exos promotes angiogenesis in diabetic wound healing via the S1PR1/AKT/FN1 signaling pathway. Burns Trauma. 2023;11:tkad003. doi:10.1093/burnst/tkad003
38. Chen X, Yao N, Mao Y, et al. Activation of the Wnt/β-catenin/CYP1B1 pathway alleviates oxidative stress and protects the blood-brain barrier under cerebral ischemia/reperfusion conditions. Neural Regen Res. 2024;19(7):1541–1547. doi:10.4103/1673-5374.386398
39. Yang Y, Liu H, Hu J, et al. Erxian decoction ameliorates myocardial tissue damage through activating PI3K/AKT signaling pathway in ovariectomized rats. Ann Med. 2024;56(1):2411013. doi:10.1080/07853890.2024.2411013
40. Rong R, Xijun X. Erythropoietin pretreatment suppresses inflammation by activating the PI3K/Akt signaling pathway in myocardial ischemia-reperfusion injury. Exp Ther Med. 2015;10(2):413–418. doi:10.3892/etm.2015.2534
41. Cui H, Zhang Q. Dexmedetomidine ameliorates lipopolysaccharide-induced acute lung injury by inhibiting the PI3K/Akt/FoxO1 signaling pathway. J Anesth. 2021;35(3):394–404. doi:10.1007/s00540-021-02909-9
42. Zhang W, Zhang JQ, Meng FM, Xue FS. Dexmedetomidine protects against lung ischemia-reperfusion injury by the PI3K/Akt/HIF-1α signaling pathway. J Anesth. 2016;30(5):826–833. doi:10.1007/s00540-016-2214-1
43. Ha DP, Lee AS. Insulin-like growth factor 1-receptor signaling stimulates GRP78 expression through the PI3K/AKT/mTOR/ATF4 axis. Cell Signal. 2020;75:109736. doi:10.1016/j.cellsig.2020.109736
44. Deng X, He Y, Miao X, Yu B. ATF4-mediated histone deacetylase HDAC1 promotes the progression of acute pancreatitis. Cell Death Dis. 2021;12(1):5. doi:10.1038/s41419-020-03296-x
45. Bai AP. β2-glycoprotein I and its antibodies involve in the pathogenesis of the antiphospholipid syndrome. Immunol Lett. 2017;186:15–19. doi:10.1016/j.imlet.2017.03.013
46. Tu HY, Li YL. Inflammation balance in skeletal muscle damage and repair. Front Immunol. 2023;14. doi:10.3389/fimmu.2023.1133355
47. den Hengst WA, Gielis JF, Lin JY, et al. Lung ischemia-reperfusion injury: a molecular and clinical view on a complex pathophysiological process. Am J Physiol Heart Circ Physiol. 2010;299(5):H1283–H1299. doi:10.1152/ajpheart.00251.2010
48. Ta HQ, Kuppusamy M, Sonkusare SK, et al. The endothelium: gatekeeper to lung ischemia-reperfusion injury. Respir Res. 2024;25(1):172. doi:10.1186/s12931-024-02776-4
49. Lim SH, Song KS, Lee J. Butyrate and propionate, short chain fatty acids, attenuate myocardial damages by inhibition of apoptosis in a rat model of ischemia-reperfusion. J Korean Soc Appl Biol Chem. 2010;53(5):570–577. doi:10.3839/jksabc.2010.088
50. Liu Y, Yang H, Zhu F, et al. Inhibition of STAT3 phosphorylation by colchicine regulates NLRP3 activation to alleviate sepsis-induced acute lung injury. Inflammopharmacology. 2023;31(4):2007–2021. doi:10.1007/s10787-023-01199-9
51. Zhang F, Hu L, Wu YX, et al. Doxycycline alleviates paraquat-induced acute lung injury by inhibiting neutrophil-derived matrix metalloproteinase 9. Int Immunopharmacol. 2019;72:243–251. doi:10.1016/j.intimp.2019.04.015
52. Lange PS, Chavez JC, Pinto JT, et al. ATF4 is an oxidative stress-inducible, prodeath transcription factor in neurons in vitro and in vivo. J Exp Med. 2008;205(5):1227–1242. doi:10.1084/jem.20071460
53. Lv Z, Liu P, Yang Y, et al. (-)-Epicatechin regulates endoplasmic reticulum stress and promotes ferroptosis in lung cancer cells via the PERK/eIF2α/ATF4 signaling pathway. PLoS One. 2024;19(10):e0313010. doi:10.1371/journal.pone.0313010
54. Chen X, Wang JH, Gao XL, et al. Tauroursodeoxycholic acid prevents ER stress-induced apoptosis and improves cerebral and vascular function in mice subjected to subarachnoid hemorrhage. Brain Res. 2020:1727. doi:10.1016/j.brainres.2019.146566
55. Tang S, Wu X, Dai Q, et al. Vitamin D receptor attenuates ischemia-reperfusion kidney injury via inhibiting ATF4. Cell Death Discov. 2023;9(1):158. doi:10.1038/s41420-023-01456-4
56. Li R, Yang LB, Li SL, et al. C/EBPα alleviates hepatic ischemia-reperfusion injury by inhibiting endoplasmic reticulum stress via HDAC1-mediated deacetylation of ATF4. J Biochem mol Toxicol. 2024;38(1). doi:10.1002/jbt.23630
57. Satyananda V, Oshi M, Tokumaru Y, et al. Sphingosine 1-phosphate (S1P) produced by sphingosine kinase 1 (SphK1) and exported via ABCC1 is related to hepatocellular carcinoma (HCC) progression. Am J Cancer Res. 2021;11(9):4394–4407.
58. Zeng M, Feng A, Wang L, et al. Aralia saponin A isolated from achyranthes bidentata bl. ameliorates LPS/D-GalN induced acute liver injury via SPHK1/S1P/S1PR1 pathway in vivo and in vitro. Int Immunopharmacol. 2023;124(Pt A):110912. doi:10.1016/j.intimp.2023.110912
59. Münzer P, Schmid E, Walker B, et al. Sphingosine kinase 1 (Sphk1) negatively regulates platelet activation and thrombus formation. Am J Physiol Cell Physiol. 2014;307(10):C920–C927. doi:10.1152/ajpcell.00029.2014
60. Aoki M, Aoki H, Mukhopadhyay P, et al. Sphingosine-1-Phosphate facilitates skin wound healing by increasing angiogenesis and inflammatory cell recruitment with less scar formation. Int J mol Sci. 2019;20(14):3381. doi:10.3390/ijms20143381
61. Li F, Yan W, Dong W, et al. PNSC928, a plant-derived compound, specifically disrupts CtBP2-p300 interaction and reduces inflammation in mice with acute respiratory distress syndrome. Biol Direct. 2024;19(1):48. doi:10.1186/s13062-024-00491-0
62. Wang Y, Mi Y, Tian J, et al. Intermedin alleviates renal ischemia-reperfusion injury and enhances neovascularization in wistar rats. Drug Des Devel Ther. 2020;14:4825–4834. doi:10.2147/dddt.S253019
63. Jin Y, Zhang M, Li M, et al. SIX1 activation is involved in cell proliferation, migration, and anti-inflammation of acute ischemia/reperfusion injury in mice. Front Mol Biosci. 2021;8:725319. doi:10.3389/fmolb.2021.725319
64. Zhou P, Song NC, Zheng ZK, et al. MMP2 and MMP9 contribute to lung ischemia-reperfusion injury via promoting pyroptosis in mice. BMC Pulm Med. 2022;22(1). doi:10.1186/s12890-022-02018-7
65. Jung HJ, Park SH, Cho KM, et al. Threonyl-tRNA synthetase promotes T helper type 1 cell responses by inducing dendritic cell maturation and IL-12 production via an NF-κB pathway. Front Immunol. 2020;11:571959. doi:10.3389/fimmu.2020.571959
66. Yang Y, Ren PW, Liu XF, et al. PPP1R26 drives hepatocellular carcinoma progression by controlling glycolysis and epithelial-mesenchymal transition. J Exp Clin Cancer Res. 2022;41(1). doi:10.1186/s13046-022-02302-8
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
Recommended articles
Analysis of Key Genes for Slow Transit Constipation Based on RNA Sequencing
Yu L, Yang X, Guan W, Zhang D, Ren S, Xing Y, An D, Zhang J, Zhu Y, Zhu A
International Journal of General Medicine 2022, 15:7569-7579
Published Date: 28 September 2022