Back to Journals » Journal of Inflammation Research » Volume 17
Identifying the ceRNA Regulatory Network in Early-Stage Acute Pancreatitis and Investigating the Therapeutic Potential of NEAT1 in Mouse Models
Received 6 August 2024
Accepted for publication 24 October 2024
Published 2 November 2024 Volume 2024:17 Pages 8099—8115
DOI https://doi.org/10.2147/JIR.S490315
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Adam Bachstetter
Bi Lin,1 Chaohao Huang2
1Department of Anesthesiology, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, People’s Republic of China; 2Department of Hepatological Surgery, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, People’s Republic of China
Correspondence: Chaohao Huang, Department of Hepatological Surgery, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, 325000, People’s Republic of China, Email [email protected]
Purpose: Acute pancreatitis (AP) is a common digestive disorder characterized by high morbidity and mortality. This study aims to uncover differentially expressed long noncoding RNAs (lncRNAs) and mRNAs, as well as related pathways, in the early stage of acute pancreatitis (AP), with a focus on the role of Neat1 in AP and severe acute pancreatitis (SAP).
Methods: In this study, we performed high-throughput RNA sequencing on pancreatic tissue samples from three normal mice and three mice with cerulein-induced AP to describe and analyze the expression profiles of long non-coding RNAs (lncRNAs) and mRNAs. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were conducted on the differentially expressed mRNAs to identify enriched pathways and biological processes. An lncRNA-miRNA-mRNA interaction network was constructed to elucidate potential regulatory mechanisms. Furthermore, we utilized Neat1 knockout mice to investigate the role of Neat1 in the pathogenesis of cerulein-AP and L-arginine-severe acute pancreatitis (SAP).
Results: Our results revealed that 261 lncRNAs and 1522 mRNAs were differentially expressed in the cerulein-AP group compared to the control group. GO and KEGG analyses of the differentially expressed mRNAs indicated that the functions of the corresponding genes are enriched in cellular metabolism, intercellular structure, and positive regulation of inflammation, which are closely related to the central events in the pathogenesis of AP. A ceRNA network involving 5 lncRNAs, 226 mRNAs, and 61 miRNAs were constructed. Neat1 was identified to have the potential therapeutic effects in AP. Neat1 knockout in mice inhibited pyroptosis in both the AP/SAP mouse models.
Conclusion: We found that lncRNAs, particularly Neat1, play a significant role in the pathogenesis of AP. This finding may provide new insights into further exploring the pathogenesis of SAP and could lead to the identification of new targets for the treatment of AP and SAP.
Keywords: acute pancreatitis, lncRNA, ceRNA, NEAT1, pyroptosis
Introduction
Acute pancreatitis (AP) is a disease characterized by the pancreas essentially digesting itself, triggered by the activation of trypsin due to a variety of causes.1 It is a significant health concern, being one of the primary reasons for hospital admissions related to gastrointestinal issues.2,3 Of those diagnosed with AP, about 20% may experience a progression to severe acute pancreatitis (SAP), which is accompanied by local or systemic complications and often requires invasive interventions. This can drive the mortality rate for SAP as high as 30%.4,5 Current therapies for acute pancreatitis (AP) are largely non-specific and supportive in nature, a necessity given the disease’s potential for rapid and unpredictable fluctuations in severity and clinical progression.6 Hence, it is crucial to explore these molecular mechanisms and identify potential biomarkers and novel therapeutic targets.
Accumulating body of research indicates that non-coding RNAs (ncRNAs), such as long noncoding RNAs (lncRNAs), circular RNAs (circRNAs), and microRNAs (miRNAs), play a significant role in the onset and progression of numerous diseases. These include lung cancer,7 inflammatory bowel disease,8 cancer resistance,9 cardiovascular disease10 and pancreatic disorders, among others. Furthermore, the expression of ncRNAs is tightly regulated under normal physiological conditions.11 Therefore, altered expression of ncRNAs could potentially serve as diagnostic biomarkers or therapeutic targets for diseases. Studies have revealed that certain lncRNAs are implicated in the pathophysiological mechanisms of acute pancreatitis (AP), participating in various processes such as the regulation of macrophage polarization,12 the preservation of gastrointestinal motility dysfunction,13 and and the induction of pancreatitis-associated lung injury.14 For example, during the inflammatory response in AP, lncRNAs primarily modulate the NF-κB pathway either directly or indirectly, triggering the expression and secretion of inflammatory factors and promoting M1 macrophage polarization. LncRNA Fendrr directly binds to the ANXA2 protein to facilitate acinar cell apoptosis. ANXA2, predominantly located in the nucleus, can attach to the 3’-UTRs of specific mRNAs to suppress their translation.15 The transcription factor CEBPB can bind to the promoter region of lncRNA MALAT1 and enhance its expression. LncRNA MALAT1 then interacts with CIRBP and prevents its ubiquitination, leading to the activation of the ERK pathway and subsequent gastrointestinal motility disorders.13 However, the role of lncRNAs in the early stages of acute pancreatitis (AP) has not been subject to comprehensive analysis.
As reported, lncRNAs have been shown to interact with a variety of targets, governing cellular functions and the expression of downstream genes.16 Among the mechanisms by which lncRNAs exert their regulatory effects, competing endogenous RNA (ceRNA) is a significant pathway. It involves lncRNAs interacting with microRNAs (miRNAs) to modulate the expression of downstream genes.17 Among the mechanisms by which lncRNAs exert their regulatory effects, competing endogenous RNA (ceRNA) is a significant pathway. It involves lncRNAs interacting with microRNAs (miRNAs) to modulate the expression of downstream genes.18 For instance, the lncRNA TCONS_00021785 elevates the expression of the E3 ligase enzyme Trim33 by sequestering miR-21-5p, thereby modulating VMP1-mediated zymophagy and diminishing the activation of trypsinogen.19 LncRNAs NONRATT022624 and NONRATT031002 boost the transcription factor ER1’s expression by interacting with miR-214-3p and miR-764-5p, respectively, which in turn regulates tissue factor expression and intensifies trypsinogen activation.20 Regarding the influence on autophagy’s onset, lncRNA PVT1 stimulates beclin1 expression through binding to miR-30a-5p, leading to abnormal autophagy induction, while lncRNA FENDRR represses ATG7 expression by associating with the RNA-binding protein PRC2, which interacts with the ATG7 promoter regions.21 The long nonprotein-coding RNA NEAT1, which associates with forming paraspeckles and maintaining their integrity, is implicated in the development and progression of a variety of cancers22–24 and inflammation.25–27 In recent years, NEAT1 has been found to play critical roles in the molecular mechanism, diagnosis, and therapy of AP. But the exact mechanism and role in the progression of pancreatitis are still unclear.
In this study, we employed RNA sequencing technology to analyze the expression profiles of the transcriptome during the initial stages of acute pancreatitis in a mouse model and inferred their functional associations through Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. Furthermore, we constructed a competing endogenous RNA (ceRNA) network to elucidate the interactions between lncRNAs and mRNAs. Additionally, we verified that NEAT1 acts as a candidate therapeutic target by regulating RHOB/NLRP3/GSDMD-dependent pyroptosis during the progression of acute pancreatitis, thereby providing a novel theoretical foundation for potential clinical treatment strategies and early intervention in acute pancreatitis.
Materials and Methods
Mice and Reagents
The C57BL/6 strain of mice, which are of the wild-type variety, were procured from Vital River Laboratory Animal Technology in Beijing, China. Simultaneously, Neat1 knockout mice (Neat−/−) were obtained from the Model Animal Research Center located in Nanjing, China. These mice were accommodated in a pathogen-free environment at Wenzhou Medical University (WMU), maintained at a constant temperature of 25°C with a lighting regimen of 12 hours of light followed by 12 hours of darkness. For the acute pancreatitis experiments, male mice aged between 8 to 10 weeks were chosen. All experiments involving mice adhered to the ethical guidelines provided by the US National Institutes of Health for laboratory animal care and use, and all protocols were authorized by the Scientific Investigation Board of Wenzhou Medical University (Approval id: wydw2022-0566). Primary antibodies to the following proteins were used in this study: GAPDH (36KD; 1:3000; 1E6D9; Proteintech Group Inc)., RhoB (22KD; 1:1000; ab277779; Abcam), NLRP3 (118KD; 1:1000; ab263899; Abcam), and GSDMD (53KD; 1:1000; ab209845; Abcam), MPO (1:50; ab9535; Abcam), Caspase-1 (48KD; 1:1000; 83383; Cell Signaling Technology) and cleaved-Caspase-1 (22KD; 1:1000; 89332; Cell Signaling Technology). The secondary antibodies are HRP-conjugated Goat Anti-Rabbit IgG (H+L) (1:3000;SA00001-2; Proteintech Group Inc). and HRP-conjugated Goat Anti-Mouse IgG (H+L) (1:3000;SA00001-1; Proteintech Group Inc). L-Arginine was sourced from Sigma-Aldrich (St. Louis, MO, USA), and Cerulein was obtained from MedChemExpress (Monmouth Junction, NJ, USA).
Experimental AP Mice Model
Before inducing experimental AP or SAP, 8-week-old wild-type mice and age-matched Neat1 knockout mice were subjected to an 8-hour fasting period. Subsequently, the mice were randomly and blindly divided into the indicated groups (n = 3 mice per group) and treated with 200 ul of cerulein at a dose of 200 ug/kg/h, administered intraperitoneally in 10 doses, or with L-Arginine at a dose of 2.5 g/kg, administered intraperitoneally in two doses with a 1-hour interval between doses, as previously described.28–30 The mice were euthanized using carbon dioxide (CO2) inhalation at either 1 or 12 hours following the initial AP induction. Serum and pancreatic tissue were then harvested for subsequent analysis. For histological examination, the pancreases were immersion-fixed in 4% paraformaldehyde, while for RNA extraction, they were preserved in RNAlater (QIAGEN, Düsseldorf, Germany). Moreover, freshly excised pancreases were rapidly frozen for later protein extraction and Western blot analysis.
Enzymatic Method Measurement of Serum Amylase and Lipase
Blood was collected from the acute pancreatitis mouse models and allowed to clot naturally at room temperature for 30 minutes. Serum was then separated through centrifugation at 4000 rpm, corresponding to a force of 1500 g, for a duration of 10 minutes at a temperature of 4 °C. The obtained serum samples were subsequently diluted to the necessary concentration and mixed with the reagents included in the assay kits. The activity levels of amylase and lipase were measured using the α-Amylase Assay Kit (C016-1) and Lipase Activity Kit (A054-2), respectively, in accordance with the instructions provided by the manufacturer (Nanjing Jiancheng Bioengineering Institute, Nanjing, China).
Protein Extraction and Western Blotting
Pancreatic tissues that had been snap-frozen were thawed and homogenized in a Triton X-100 lysis buffer supplemented with a complete protease inhibitor cocktail (Roche), as previously described.28 The protein concentration in the lysates was quantified using a BCA Protein Assay Kit (Beyotime Biotechnology, Shanghai, China) and then diluted to equal concentrations with the lysis buffer. Equivalent amounts of protein were separated by SDS-PAGE, transferred to PVDF membranes (Millipore). The membranes were blocked with 5% skimmed milk and then incubated with primary antibodies mentioned above overnight at 4 °C and were incubated with a HRP-conjugated Goat Anti-Rabbit IgG (H+L) or HRP-conjugated Goat Anti-Mouse IgG (H+L) at room temperature for 1 h. After the membrane is washed with eluent, the immunoreactive bands were visualized using SuperFemto ECL Chemiluminescence Kit (E423-01; Vazyme) and then blotted with the iBright FL Imaging System (Thermo Fisher Scientific, Waltham, MA, USA). The intensity of WB band can be calculated by imageJ software. GAPDH was utilized as a loading control.
Pancreas Histological Examination, Immunohistochemistry and Detection of Myeloperoxidase (MPO) Levels
Twelve hours following the induction of acute pancreatitis (AP), pancreatic specimens were harvested and fixed in a 4% paraformaldehyde solution in PBS. Subsequently, the fixed tissues were embedded in paraffin wax, sectioned into 5-μm slices, and subjected to staining with hematoxylin and eosin (H&E), an antibody against myeloperoxidase (MPO) from Abcam (1:50; ab9535), and a TUNEL assay kit (C1091) from Beyotime Biotechnology (Shanghai, China), following previously established protocols.31
Pancreatic tissue samples were thawed and homogenized in 20 mm phosphate-buffered saline (PBS) at pH 7.4 using a tissue homogenizer. The homogenate was then centrifuged at 1200 g for 20 minutes at 4 °C. The supernatant was collected and analyzed for MPO activity using an enzyme-linked immunosorbent assay (ELISA) kit (EMMPO, Thermo Fisher Scientific), following the manufacturer’s instructions.
RNA Isolation and Quantitative PCR (qPCR)
RNA was isolated from the pancreatic tissue stored in RNAlater using the TRIzol reagent (ThermoFisher Scientific), in accordance with the manufacturer’s instructions. Then, reverse transcription for mRNA detection was performed using PrimeScript™ RT Reagent Kit for RT-PCR (Takara Shuzo Co., Tokyo), in line with the kit’s recommended procedures. And reverse transcription for miRNA, U6 snRNA, was performed using the miRNA 1st-Strand cDNA Synthesis Kit (Vazyme, MR201-01, Nanjing, China). Following cDNA synthesis, real-time PCR was performed on a Bio-Rad CFX instrument, utilizing the SYBR Green reagent provided by TAKARA Biotech (Tokyo, Japan). The relative expression of the genes of interest was normalized to the housekeeping gene Rpl32 or U6, using the 2^-ΔΔCt method for cycle threshold analysis.28 The primer sequences for qPCR were list in Table 1.
 |
Table 1 Sequences of the Primers Used for Quantitative Real-Time PCR |
RNA-Sequencing Data Acquisition, Quality Control, and Processing
RNA was isolated from the whole pancreas of cerulein-induced acute pancreatitis (AP) mouse models. RNA concentration and quality were determined using a Qubit 2.0 Fluorometer (Thermo Fisher) and an Agilent 2100 Bioanalyzer, respectively. Libraries were constructed with the TruSeq Stranded mRNA Sample Prep Kit (Illumina) according to the kit’s protocol. Library quantification and quality checks were performed using the Qubit 2.0 Fluorometer and the Agilent 2100 Bioanalyzer, and the library molar concentration was verified by qPCR before pooling. Sequencing was conducted on a HiSeq X10 system using a 2×150 bp paired-end, dual-index approach. For RNA-Seq data processing, Trimmomatic was used to remove adapters, trim poor-quality bases, and discard reads shorter than 36 bp. The processed reads were aligned to the mouse mm10 genome using the STAR aligner. Gene read counts were obtained with HTSeq’s htseq-count command. Standardized read counts were used to calculate the differential expression of lncRNAs (DEL) and mRNAs (DEM) between the two groups. In order to adjust for multiple comparisons and control for Type I error, R’s limma and edgeR were used to identify differentially expressed genes (DEGs) with a significance level of FDR ≤ 0.01 and a absolute value of log fold change (|logFC|) of ≥ 2. Cluster analysis of DEGs was performed using R’s pheatmap and ggplot2 functions with normalized expression levels. Venn diagrams, Volcano plots, and heatmaps were generated to visualize differential expression, with up-regulated and down-regulated DEGs represented by different colors on the heatmaps. For the DEGs, Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) biological process enrichment analyses were conducted using Fisher’s exact test, with p-values adjusted by the Bonferroni method. The RNA-Seq data have been deposited in the Gene Expression Omnibus (GEO accession number GSE272464).
Construction of lncRNA-miRNA-mRNA Network
LncRNA and mRNA exhibit sequence similarity, expression correlation, and positional neighbor relationship, both exerting their roles through competitive binding with miRNA molecules (Tay et al, 2014). Consequently, different RNA molecules competing for the same miRNA establish a competitive relationship. To predict the interactions between miRNAs and DEMs, DELs respectively, we employed mircode, Targetscan (Release 8.0), miRTarBase (Release 9.0) and miRDB (version 6.0) software based on base pairing principles. We integrated these predicted results to construct potential lncRNA-miRNA-mRNA ceRNA networks and visualized them using Cytoscape software (version 3.10.0) in order to explore the role of lncRNAs in SAP pathogenesis.
Statistical Analysis
Data are presented as mean ± standard deviation (SD). The data must undergo normality testing before analysis. For datasets that follow a normal distribution, a two-tailed Student’s t-test was employed for comparing between two groups, while a one-way ANOVA, followed by Tukey’s post-hoc tests was used for comparing between more than two groups. Kaplan-Meier method was used to analyze the survival rates of the wild-type and Neat1−/− mice following the administration of the lethal dose of L-Arginine. All analyses were conducted using GraphPad Prism version 10 (GraphPad Software Inc)., *p < 0.05, **p < 0.01, n.s. represents non-significant, respectively.
Results
Analysis of Differentially Expressed mRNAs, Including Gene Ontology and KEGG Pathway Assessments
To delve into the pathogenic mechanisms underlying the early phase of acute pancreatitis (AP), we conducted a comprehensive RNA-sequencing analysis to profile differentially expressed mRNAs (DEMs, Figure 1A). We deemed a significant difference to exist when the |logFC| was ≥ 2 and the FDR was <0.01, the top 50 genes were selected to plot a heatmap (Figure 1B). Our analysis revealed that 1522 mRNAs exhibited differential expression in the AP group, with 839 being upregulated and 683 downregulated when compared to the control group (Figure 1C). Visual representation of these differences was provided by a Volcano plot and hierarchical clustering analysis, which depicted the distinct mRNA expression patterns in pancreatic tissues between the control and AP groups. The PDF of partial DEMs heatmap was attached to Figure S1, and the full list of DEMs was shown in Table S1.
To elucidate the functional implications of the DEMs, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. The GO analysis of the DEMs revealed their biological process (BP) functions to be predominantly associated with small molecule catabolic processes, organic acid catabolism, and carboxylic acid catabolism. In terms of cellular components (CC), the DEMs were linked to cell-substrate junctions, focal adhesions, and the cell cortex. The molecular functions (MF) predicted for these DEMs included actin binding, cadherin binding, and virus receptor activity (Figure 1D). KEGG pathway analysis uncovered the potential pathways by which these DEMs might influence AP pathogenesis, highlighting their involvement in the regulation of cytokine-cytokine receptor interactions, the inflammatory signaling pathway, and the cytoskeleton in muscle cells (Figure 1E).
Differential Expression of lncRNAs and Construction of lncRNA-miRNA-mRNA Network
The RNA-Seq analysis was employed in our study to examine the differentially expressed long non-coding RNAs (lncRNAs) during the quiescent and initial stages of acute pancreatitis (AP). This analysis resulted in the identification of a total of 261 differentially expressed lncRNAs. The top 50 lncRNAs, based on their expression changes, were selected for display in Figure 2A, with 139 showing upregulation and 122 showing downregulation. Hierarchical cluster analysis and a volcano plot (Figure 2B) revealed significant differences in lncRNA expression between the pancreatic tissues of the control group and the AP group. LncRNAs are known to participate in the regulation of biological processes in various ways, such as directly interacting with mRNA to alter the spatial conformation of chromatin or being influenced by regulatory molecules. The PDF of partial DELs heatmap was attached to Figure S2, and the full list of DELs was shown in Table S2.
Considering the interplay between lncRNAs and mRNAs, and the role of miRNAs as intermediary regulatory molecules, we constructed a competing endogenous RNA (ceRNA) network to predict the functions of differentially expressed lncRNAs (DELs) and differentially expressed mRNAs (DEMs). As depicted in Figure 2C, 226 genes among the DEMs were identified as potential targets regulated by the annotated DELs. Upon summarization, a ceRNA network was constructed, involving 5 lncRNAs, 226 mRNAs, and 61 miRNAs, with each differentially expressed gene being associated with one or more miRNAs (Figure 2D). The ceRNA network suggests that these lncRNAs not only independently participate in the regulation of biological processes but also act as miRNA sponges, collectively affecting downstream gene expression and contributing to the onset and progression of AP. The PDF of the ceRNA network was attached to Figure S3, and the detailed information of the ceRNA is listed in Table S3.
The ceRNA network emerges as a pivotal regulatory mechanism in the pathogenesis of AP. This could lead to the overexpression of genes that are detrimental to pancreatic acinar cell homeostasis, potentially exacerbating inflammation and tissue damage. The implications of a dysregulated ceRNA network in AP suggest that therapies targeting this network could have broad-spectrum effects. Since ceRNAs can regulate multiple mRNA targets, interventions aimed at normalizing ceRNA activity could simultaneously modulate several disease pathways, offering a more comprehensive approach to treatment than agents targeting single molecules. However, it is important to note that the complexity of ceRNA networks necessitates a nuanced understanding of their dynamics. The interplay between various ceRNA molecules and their targets is likely to be context-dependent, and further research is needed to fully elucidate the network’s role in AP. Additionally, potential off-target effects and the long-term consequences of ceRNA network modulation must be carefully evaluated.
Verify Differentially Dysregulated lncRNAs and mRNAs
Based on the result of the RNA-Seq and the constructed lncRNA-miRNA-mRNA Network, we verified some of the lncRNAs and genes we were interested in. Compared with control group, the lncRNA and genes selected in AP group, including H19, Mir17hg, Mir22hg, Jpx, Neat1, Rhob, Nlrp3, Cxcl1, Cxcl2, il1b were significantly overexpressed and consistent with the RNA Sequencing results (Figure 3A and B). Additionally, We confirmed that specific microRNAs are predicted to be involved in the ceRNA network during the early stage of AP, including mir-206-3p, mir-128-3p, mir-24-3p, mir-223-3p, mir-7-5p (Figure 3C). Among them, Neat1 was the lncRNA with the highest connectivity in the ceRNA network, which suggested that NEAT1 might be the most functional regulator and play a great role in the pathogenesis of AP. Thus, we employed the lncRNA neat1 knockout mice to explore the effects of Neat1 on AP mouse model.
LncRNA Neat1 Knockout Alleviated Cerulein-Induced AP and L-Arginine-Induced SAP in Mice
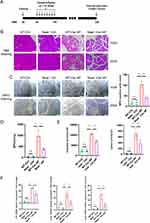
To confirm whether increased Neat1 expression mediates the exacerbation of acute pancreatitis (AP), we established a cerulein-induced AP model in wild-type (WT) and Neat1 knockout (Neat1−/−) mice (Figure 4A). Administration of cerulein resulted in dramatic pathological changes, including pancreas edema, inflammatory cell infiltration, and tissue necrosis. However, Neat1 knockout prevented these pancreatitis symptoms induced by cerulein injection (Figure 4B). Neutrophils were recruited into the injured pancreases during pancreatitis progression, whereas significantly fewer MPO+ cells [Figure 4C, ANOVA summary: F (3, 8) = 40.74, P<0.0001, R2=0.9386] and lower pancreatic MPO activity [Figure 4D, ANOVA summary: F (3, 8) = 34.34, P<0.0001, R2=0.9279] were observed in the injured pancreases from cerulein-AP mice pretreated with Neat1 knockout. Consistently, Neat1 knockout inhibited the elevation of serum amylase activity [ANOVA summary: F (3, 8) = 148.5, P<0.0001, R2=0.9824] and lipase activity [ANOVA summary: F (3, 8) = 31.29, P<0.0001, R2=0.9215] in the cerulein-AP mouse model (Figure 4E). It also suppressed the induction of inflammatory cytokine and chemokine gene expression, including il1b [ANOVA summary: F (3, 8) = 52.67, P < 0.0001, R2 =0.9518], cxcl1 [ANOVA summary: F (3, 8) = 30.33, P = 0.0001, R2 =0.9192], and cxcl2 [ANOVA summary: F (3, 8) = 20.27, P = 0.0004, R2 =0.8843], in the injured pancreases from cerulein-AP mice (Figure 4F).
We also tested the damaging effect of Neat1 in L-arginine-induced severe acute pancreatitis (SAP) mouse models (Figure 5A). Similarly, Neat1 knockout also prevented these pancreatitis symptoms (Figure 5B), neutrophil infiltration measured by MPO+ cells [Figure 5C, ANOVA summary: F (3, 8) = 33.04, P < 0.0001, R2 = 0.9253] and MPO activity [Figure 5D, ANOVA summary: F (3, 8) = 38.47, P < 0.0001, R2 = 0.9352], inhibited the elevation of serum amylase activity [ANOVA summary: F (3, 8) = 13.30, P = 0.0018, R2 = 0.8330] and lipase activity [ANOVA summary: F (3, 8) = 51.05, P < 0.0001, R2 = 0.9504] (Figure 5E), suppressed the induction of inflammatory cytokine and chemokine gene expression, including il1b [ANOVA summary: F (3, 8) = 19.34, P=0.0005, R2 =0.8788], cxcl1 [ANOVA summary: F (3, 8) = 23.44, P=0.0003, R2 =0.8978], and cxcl2 [ANOVA summary: F (3, 8) = 74.30, P < 0.0001, R2 =0.9654] (Figure 5F), in the L-arginine-SAP model. In addition, the wild-type mice were significantly more susceptible to SAP and exhibited significantly higher mortality rates compared to the Neat1−/− mice (Figure 5G, χ2 = 4.299, P = 0.038) when treated with the lethal dose of L-Arginine (4g/kg).29 These results demonstrate that loss of Neat1 within the injured pancreatic acinar cell ameliorates the severity of experimental AP.
LncRNA Neat1 Knockout Inhibited Pyroptosis in AP/SAP Mouse Models
In these differently expression mRNAs, we observed that neat1 co-increased with nlrp3, il1b and rhob. Previously studies have demonstrated that the factors mentioned above are positive associated with pyroptosis. Therefore, we aimed to explore the role of Neat1 played in AP mouse models, and whether Neat1 knockout affected the expression of pyroptosis associated proteins. As shown in Figure 6A and B, Western blotting analysis showed that the protein expression levels of Rhob, Nlrp3, Cleaved-Caspase-1, and Gsdmd were significantly upregulated in the pancreas tissues of cerulein-AP model mice and L-arginine-SAP model mice. However, Neat1 knockout significantly reversed these effects. Besides, no significant differences were observed in the protein expression levels of Caspase-1 between the AP and SAP groups. We further perform TUNEL staining to assess whether Neat1 deficiency could influence post-AP/SAP pancreas cell damage, TUNEL+ cells were significantly decreased in the pancreatic acinar cells of Neat1−/− AP [ANOVA summary: F (3, 8) = 45.76, P < 0.0001, R2 =0.9449] and Neat1−/− SAP [ANOVA summary: F (3, 8) = 104.3, P < 0.0001, R2 =0.9751] mice versus the WT AP/SAP mice (Figure 6C and D). The above results demonstrated that NEAT1 played a great role in the pathogenesis of AP/SAP.
Discussions
Acute pancreatitis (AP) is a common acute abdominal disease with high mortality and mortality rates.32 About 80% patients with AP have mild and moderate symptoms, and will recover in 1–2 weeks under the reasonable treatment. However, the remaining 20% AP patients will develop into SAP with the 5–10% overall case fatality rate.33,34 Despite the extensive accumulation of research documents dedicated to finding novel therapeutic targets and intervention checkpoints for the treatment of acute pancreatitis (AP), the results have been modest. Acute pancreatitis remains a challenging disease with limited treatment options, particularly in cases of severe acute pancreatitis (SAP).6 Consequently, further research and clinical trials are necessary to develop more effective treatments and improve patient outcomes. Screening for gene expression changes at the onset of pancreatitis may provide an opportunity to identify new therapeutic targets for pancreatitis. In this study, we employed RNA-Seq to discover the gene networks and corresponding important network nodes involved in a mouse model of AP. The sequence results demonstrated that 1522 mRNAs and 261 lncRNAs were differentially expressed, and that these differential genes were mainly highly expressed. Based on this result, the present study established a genetic interaction network in initial stage of AP. Through functional analysis, the present study demonstrated that these genes primarily play roles in regulating cellular metabolism, intercellular structure, and positive inflammation, including processes such as organic acid catabolism, carboxylic acid catabolism, cell-substrate junction formation, focal adhesion, cortical cytoskeleton organization, as well as activities like MAP kinase tyrosine phosphatase activity, MAP kinase phosphatase activity, and NAD binding. The full list of GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment pathways can be found in the supplementary material.
In our study, the gene network in the initial stage of acute pancreatitis (AP) revealed several potentially important genes, including neat1, il1b, nlrp3, and rhob, which are closely associated with the pathogenesis of AP and pyroptosis. The role of pyroptosis in acute pancreatitis is an area of intense research. Inhibition of these initially upregulated genes might suppress the pathogenesis of AP. Studies have suggested that the inflammasome and the subsequent pyroptotic process may contribute to the pathogenesis of AP. Inflammatory responses triggered by pyroptosis, such as the release of IL-1β, could exacerbate both local and systemic inflammation in AP. Furthermore, inflammasome activation and the release of pro-inflammatory cytokines can recruit immune cells, such as neutrophils, potentially intensifying the inflammatory response and causing pancreatic tissue damage.35 Research has shown that inhibiting pyroptosis by targeting the NLRP3/Caspase1/GSDMD pathway could be a novel therapeutic approach for AP.36,37 Activated RhoB has been found to promote the activation of ROCK and the formation of the NLRP3 inflammasome, facilitating the occurrence of pyroptosis.38 Additionally, NEAT1 has been observed to be upregulated in certain inflammatory conditions, including allergic rhinitis,39 acute kidney injury,40 and pancreatitis.41 It is believed to interact with other RNA molecules and proteins, potentially influencing pyroptosis in pancreatic acinar cells. Downregulation of lncRNA NEAT1 relieves cerulein-induced cell apoptosis and inflammatory injury in AR42J cells through sponging miR-365a-3p in acute pancreatitis.42 Quercetin-mediated downregulation of NEAT1, which in turn reduces miR-216b expression, inhibits cerulein-induced acute pancreatitis.41 Our RNA-Seq and gene network results suggest that NEAT1 may act as a competing endogenous RNA (ceRNA) in the initial stage of AP. NEAT1 possesses the highest abundance of potential binding sites for miRNAs in the ceRNA network, which are predicted to regulate numerous downstream mRNAs. While some microRNAs (miRNAs) have been shown to interact with NEAT1 in the mircode database, there is no definitive evidence to confirm these interactions in the ENCORI/starBase database. Consequently, we focused on exploring the effects of NEAT1 in AP pathogenesis. Using Neat1 knockout mice, we investigated the impact of Neat1 on the mouse AP process. The results showed that the knockout of Neat1 significantly inhibited pyroptosis in AP/SAP.
Pyroptosis, a distinct form of programmed cell death, differs from conventional necrosis and apoptosis. It is characterized by the formation of membrane pores and the release of IL-1β, a key effector cytokine.43 Activated caspase-1 plays a crucial role in this process by cleaving the GSDMD protein, leading to the oligomerization of the GSDMD-N amino terminus. This event triggers the formation of plasma membrane pores, facilitating the secretion of IL-1β and IL-18, and ultimately induces pyroptosis.44 This process is implicated in the pathogenesis of systemic inflammation. Herein, the expression of NLRP3, a pyroptosis-related marker protein, was upregulated in both cerulein- and L-arginine-induced AP mouse model. Knockout of Neat1 alleviated AP-related tissue damage, indicating that pyroptosis is involved in the pathogenic mechanisms of AP in vivo. As demonstrated by the tunel assay, Neat1 knockout significantly increased the survival rate of pancreatic acinar cells in pancreatitis’ tissues. These results suggest that NEAT1 induces acinar cell damage by promoting pyroptosis, suggesting that NEAT1 suppression alleviates pyroptosis in AP and SAP.
Although our research is the first time to explore the theraputic effects of NEAT1 in AP/SAP knockout mice, it also has a few drawbacks and limitations: Firstly, as mentioned above, we only studied the effect of neat1 on acute pancreatitis in mice, without further studying the specific mechanism of its regulation of AP. In the future, miRNA-Seq and CLIP-Seq should be used to pinpoint the underlying mechanism of NEAT1 in acute pancreatitis. Secondly, the Neat1-deficient mice employed in this study are global knockout. Macrophage-specific Neat1 knockout mice and pancreatic acinar cell-specific Neat1 knockout mice should be used to pinpoint the major source of Neat1 in acute pancreatitis.
Conclusion
In summary, the results of the present study suggested that Neat1 was highly expressed in AP and may affect the development of pyroptosis. Further analysis revealed that a neat1-miRNA-mRNA regulatory network may be the basis for the effects of Neat1 on AP. The results of the present study may provide novel insight for studies into the pathogenesis of AP and pyroptosis, as well as the development of strategies for the clinical treatment of AP. Future research should aim to validate these findings in larger patient cohorts and explore the clinical applicability of ceRNA network modulation. NEAT1 could be a potential prognostic marker in AP and SAP. And a drug or adenovirus target to NEAT1 in pancreatic acinar cells need to be developed to treat AP/SAP in mice in our further experiments or patients.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon request. The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE272464.
Ethics Approval and Informed Consent
All animal procedures were conducted in accordance with the guidelines approved by the Institutional Animal Care and Use Committee of the Wenzhou Medical University (Approval id: wydw2022-0566).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the PhD Start-up Funding of the First Affiliated Hospital of Wenzhou Medical University, Zhejiang, China (No. 2019QD004), and Wenzhou technology bureau project(Y20210934).
Disclosure
The authors have no conflicts of interest to declare for this work.
References
1. Habtezion A, Gukovskaya AS, Pandol SJ. Acute pancreatitis: a multifaceted set of organelle and cellular interactions. Gastroenterology. 2019;156(7):1941–1950. doi:10.1053/j.gastro.2018.11.082
2. Iannuzzi JP, King JA, Leong JH, et al. Global incidence of acute pancreatitis is increasing over time: a systematic review and meta-analysis. Gastroenterology. 2022;162(1):122–134. doi:10.1053/j.gastro.2021.09.043
3. Wadhwa V, Patwardhan S, Garg SK, Jobanputra Y, Lopez R, Sanaka MR. Health care utilization and costs associated with acute pancreatitis. Pancreas. 2017;46(3):410–415. doi:10.1097/MPA.0000000000000755
4. Liu X, Luo W, Chen J, et al. USP25 deficiency exacerbates acute pancreatitis via up-regulating TBK1-NF-kappaB signaling in macrophages. Cell Mol Gastroenterol Hepatol. 2022;14(5):1103–1122. doi:10.1016/j.jcmgh.2022.07.013
5. Trikudanathan G, Wolbrink DRJ, van Santvoort HC, et al. Current concepts in severe acute and necrotizing pancreatitis: an evidence-based approach. Gastroenterology. 2019;156(7):1994–2007e1993. doi:10.1053/j.gastro.2019.01.269
6. Zhou Y, Huang X, Jin Y, et al. The role of mitochondrial damage-associated molecular patterns in acute pancreatitis. Biomed Pharmacother. 2024;175:116690. doi:10.1016/j.biopha.2024.116690
7. Yang Q, Wang W, Cheng D, et al. Non-coding RNA in exosomes: regulating bone metastasis of lung cancer and its clinical application prospect. Transl Oncol. 2024;46:102002. doi:10.1016/j.tranon.2024.102002
8. Heydari R, Karimi P, Meyfour A. Long non-coding RNAs as pathophysiological regulators, therapeutic targets and novel extracellular vesicle biomarkers for the diagnosis of inflammatory bowel disease. Biomed Pharmacother. 2024;176:116868.
9. Tufail M, Hu JJ, Liang J, et al. Hallmarks of cancer resistance. iScience. 2024;27(6):109979. doi:10.1016/j.isci.2024.109979
10. Chen DX, Lu CH, Na N, Yin RX, Huang F. Endothelial progenitor cell-derived extracellular vesicles: the world of potential prospects for the treatment of cardiovascular diseases. Cell Biosci. 2024;14(1):72. doi:10.1186/s13578-024-01255-z
11. Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96(4):1297–1325. doi:10.1152/physrev.00041.2015
12. Chen S, Zhu J, Sun LQ, et al. LincRNA-EPS alleviates severe acute pancreatitis by suppressing HMGB1-triggered inflammation in pancreatic macrophages. Immunology. 2021;163(2):201–219. doi:10.1111/imm.13313
13. Lai L, Wang G, Xu L, Fu Y. CEBPB promotes gastrointestinal motility dysfunction after severe acute pancreatitis via the MALAT1/CIRBP/ERK axis. Mol Immunol. 2023;156:1–9. doi:10.1016/j.molimm.2023.02.001
14. Xu C, Luo Y, Ntim M, et al. Effect of emodin on long non-coding RNA-mRNA networks in rats with severe acute pancreatitis-induced acute lung injury. J Cell Mol Med. 2021;25(4):1851–1866. doi:10.1111/jcmm.15525
15. Zhao D, Ge H, Ma B, et al. The interaction between ANXA2 and lncRNA Fendrr promotes cell apoptosis in caerulein-induced acute pancreatitis. J Cell Biochem. 2019;120(5):8160–8168. doi:10.1002/jcb.28097
16. Zhang Y, Liu H, Niu M, et al. Roles of long noncoding RNAs in human inflammatory diseases. Cell Death Discov. 2024;10(1):235. doi:10.1038/s41420-024-02002-6
17. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi:10.1038/nature12986
18. Song TJ, Ke J, Chen F, Zhang JY, Zhang C, Chen HY. Effect of SNHG11/miR-7-5p/PLCB1 axis on acute pancreatitis through inhibiting p38MAPK pathway. Cells. 2022;12(1):65. doi:10.3390/cells12010065
19. Wang Q, Yu J, Gao W, et al. The lncRNA TCONS_00021785/miR-21-5p/Trim33 axis regulates VMP1-mediated zymophagy, reduces the activation of trypsinogen, and promotes acinar cell recovery. Cell Death Discov. 2022;8(1):65. doi:10.1038/s41420-022-00862-4
20. Gao B, Zhang X, Xue D, Zhang W. Effects of Egr1 on pancreatic acinar intracellular trypsinogen activation and the associated ceRNA network. Mol Med Rep. 2020;22(3):2496–2506. doi:10.3892/mmr.2020.11316
21. Hu F, Tao X, Zhao L, et al. LncRNA-PVT1 aggravates severe acute pancreatitis by promoting autophagy via the miR-30a-5p/Beclin-1 axis. Am J Transl Res. 2020;12(9):5551–5562.
22. Luo X, Wei Q, Jiang X, et al. CSTF3 contributes to platinum resistance in ovarian cancer through alternative polyadenylation of lncRNA NEAT1 and generating the short isoform NEAT1_1. Cell Death Dis. 2024;15(6):432. doi:10.1038/s41419-024-06816-1
23. Gu L, Yue X, Niu S, et al. Systematical identification of key genes and regulatory genetic variants associated with prognosis of esophageal squamous cell carcinoma. Mol, Carcinog. 2024;63(6):1013–1023. doi:10.1002/mc.23704
24. Bhattacharya A, Wang K, Penailillo J, et al. MUC1-C regulates NEAT1 lncRNA expression and paraspeckle formation in cancer progression. Oncogene. 2024;43(28):2199–2214. doi:10.1038/s41388-024-03068-3
25. Ma T, Li H, Liu H, et al. Neat1 promotes acute kidney injury to chronic kidney disease by facilitating tubular epithelial cells apoptosis via sequestering miR-129-5p. Mol Ther. 2022;30(10):3313–3332. doi:10.1016/j.ymthe.2022.05.019
26. Tayel SI, El-Masry EA, Abdelaal GA, Shehab-Eldeen S, Essa A, Muharram NM. Interplay of LncRNAs NEAT1 and TUG1 in incidence of cytokine storm in appraisal of COVID-19 infection. Int J Biol Sci. 2022;18(13):4901–4913. doi:10.7150/ijbs.72318
27. Wu Y, Li P, Liu L, et al. lncRNA Neat1 regulates neuronal dysfunction post-sepsis via stabilization of hemoglobin subunit beta. Mol Ther. 2022;30(7):2618–2632. doi:10.1016/j.ymthe.2022.03.011
28. Huang C, Chen S, Zhang T, et al. TLR3 ligand polyi: C prevents acute pancreatitis through the interferon-beta/interferon-alpha/beta receptor signaling pathway in a caerulein-induced pancreatitis mouse model. Front Immunol. 2019;10:980. doi:10.3389/fimmu.2019.00980
29. Kang R, Zhang Q, Hou W, et al. Intracellular Hmgb1 inhibits inflammatory nucleosome release and limits acute pancreatitis in mice. Gastroenterology. 2014;146(4):1097–1107. doi:10.1053/j.gastro.2013.12.015
30. Yang L, Ye F, Liu J, Klionsky DJ, Tang D, Kang R. Extracellular SQSTM1 exacerbates acute pancreatitis by activating autophagy-dependent ferroptosis. Autophagy. 2023;19(6):1733–1744. doi:10.1080/15548627.2022.2152209
31. Ergashev A, Shi F, Liu Z, et al. KAN0438757, a novel PFKFB3 inhibitor, prevent the progression of severe acute pancreatitis via the Nrf2/HO-1 pathway in infiltrated macrophage. Free Radic Biol Med. 2024;210:130–145. doi:10.1016/j.freeradbiomed.2023.11.010
32. Lee PJ, Papachristou GI. New insights into acute pancreatitis. Nat Rev Gastroenterol Hepatol. 2019;16(8):479–496. doi:10.1038/s41575-019-0158-2
33. Garg PK, Singh VP. Organ Failure Due to Systemic Injury in Acute Pancreatitis. Gastroenterology. 2019;156(7):2008–2023. doi:10.1053/j.gastro.2018.12.041
34. Grasso D, Ropolo A, Lo Re A, et al. Zymophagy, a novel selective autophagy pathway mediated by VMP1-USP9x-p62, prevents pancreatic cell death. J Biol Chem. 2011;286(10):8308–8324. doi:10.1074/jbc.M110.197301
35. Xu Q, Wang M, Guo H, et al. Emodin alleviates severe acute pancreatitis-associated acute lung injury by inhibiting the cold-inducible RNA-binding protein (CIRP)-mediated activation of the NLRP3/IL-1beta/CXCL1 signaling. Front Pharmacol. 2021;12:655372. doi:10.3389/fphar.2021.655372
36. Shao Y, Jiang Y, Wang J, Li H, Li C, Zhang D. Inhibition of circulating exosomes release with GW4869 mitigates severe acute pancreatitis-stimulated intestinal barrier damage through suppressing NLRP3 inflammasome-mediated pyroptosis. Int Immunopharmacol. 2024;126:111301. doi:10.1016/j.intimp.2023.111301
37. Lu Y, Li B, Wei M, et al. HDL inhibits pancreatic acinar cell NLRP3 inflammasome activation and protect against acinar cell pyroptosis in acute pancreatitis. Int Immunopharmacol. 2023;125(Pt A):110950. doi:10.1016/j.intimp.2023.110950
38. Dong W, Liao R, Weng J, et al. USF2 activates RhoB/ROCK pathway by transcriptional inhibition of miR-206 to promote pyroptosis in septic cardiomyocytes. Mol Cell Biochem. 2024;479(5):1093–1108. doi:10.1007/s11010-023-04781-5
39. Liu Y, Gao J, Xu Q, et al. Long non-coding RNA NEAT1 exacerbates NLRP3-mediated pyroptosis in allergic rhinitis through regulating the PTBP1/FOXP1 cascade. Int Immunopharmacol. 2024;137:112337. doi:10.1016/j.intimp.2024.112337
40. Xue R, Yiu WH, Chan KW, et al. Long non-coding RNA Neat1, NLRP3 inflammasome, and acute kidney injury. J Am Soc Nephrol. 2024;35(8):998–1015. doi:10.1681/ASN.0000000000000362
41. Sheng B, Zhao L, Zang X, et al. Quercetin inhibits caerulein-induced acute pancreatitis through regulating miR-216b by targeting MAP2K6 and NEAT1. Inflammopharmacology. 2021;29(2):549–559. doi:10.1007/s10787-020-00767-7
42. Shao A, Hu W, Li C, Yang Y, Zhu J. Downregulation of lncRNA NEAT1 relieves caerulein-induced cell apoptosis and inflammatory Injury in AR42J cells through sponging miR-365a-3p in acute pancreatitis. Biochem Genet. 2022;60(6):2286–2298. doi:10.1007/s10528-022-10219-2
43. Yu P, Zhang X, Liu N, Tang L, Peng C, Chen X. Pyroptosis: mechanisms and diseases. Signal Transduct Target Ther. 2021;6(1):128. doi:10.1038/s41392-021-00507-5
44. He WT, Wan H, Hu L, et al. Gasdermin D is an executor of pyroptosis and required for interleukin-1beta secretion. Cell Res. 2015;25(12):1285–1298. doi:10.1038/cr.2015.139
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.