Back to Journals » Journal of Inflammation Research » Volume 18
IFNα/JAK/STAT1 Axis-Induced FBXO4 Modulates Muscle Cell Differentiation via β-Catenin Degradation in Dermatomyositis
Authors Yin L, Yang H, Fu M, Bai Y, Hu N, Sun H
Received 15 November 2024
Accepted for publication 28 May 2025
Published 27 June 2025 Volume 2025:18 Pages 8527—8539
DOI https://doi.org/10.2147/JIR.S506056
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Liguo Yin,1,2 Hanbo Yang,3 Min Fu,1,2 Yanyan Bai,1,2 Naiwen Hu,1,2 Hongsheng Sun1,2
1Department of Rheumatology and Immunology, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, People’s Republic of China; 2Department of Rheumatology and Immunology, Shandong Provincial Hospital, Cheeloo College of Medicine, Shandong University, Jinan, People’s Republic of China; 3Department of Rheumatology, China-Japan Friendship Hospital, Beijing, People’s Republic of China
Correspondence: Hongsheng Sun, Email [email protected]
Purpose: Dermatomyositis (DM) is an inflammatory myopathy characterized by chronic muscle inflammation and damage. Although the pathogenesis of DM has been widely reported to be related to chronic inflammation, the role of ubiquitin E3 ligases in DM remains unclear. In the current study, we aimed to investigate the biological roles of ubiquitin E3 ligase in DM.
Methods: Deseq2 was used to screen the differential express genes in DM public datasets. Quantitative real time PCR and Western blot were used to examine the mRNA and protein levels. Co-immunoprecipitation assays were used to investigate the protein interactions between proteins. Dual-luciferase reporter assays were applied to investigate the regulation between transcription factors and targets.
Results: In the current study, we screened public DM-related datasets and focused on ubiquitin-proteasome-related enzymes. Ultimately, we identified the ubiquitin E3 ligase FBXO4. FBXO4 was significantly upregulated in DM muscle tissues compared to normal controls. In human muscle cells (LHCN-M2), FBXO4 knockout led to significant upregulation of genes related to muscle cell differentiation and significant downregulation of genes enriched in cell cycle pathways, as revealed by RNA-seq. These results suggest that FBXO4 knockout promotes muscle cell differentiation. Mechanistic studies showed that FBXO4 ubiquitinates and degrades β-catenin, thereby inhibiting the Wnt/β-catenin signaling pathway and suppressing muscle cell differentiation. On the other hand, FBXO4 may promote muscle cell apoptosis in DM by degrading MCL1. Additionally, we found that FBXO4 is regulated by the IFNα/JAK/STAT1 signaling pathway in DM and identified FBXO4 as a direct target of STAT1.
Conclusion: In conclusion, our findings suggest that IFNα/JAK/STAT1 signaling pathway elevates the expression of FBXO4 in DM and then it contributes to muscle atrophy by inhibiting differentiation and promoting apoptosis. Targeting FBXO4 may offer a novel therapeutic approach for DM.
Keywords: dermatomyositis, IFNα/JAK/STAT1 axis, FBXO4, Wnt/β-catenin signaling, muscle cell differentiation
Graphical Abstract:

Introduction
Dermatomyositis (DM) is a type of inflammatory myopathy marked by distinctive skin manifestations, proximal muscle weakness, and perifascicular atrophy (PFA) detected in muscle biopsy specimens.1–3
The pathogenesis of DM is complex, involving both immune-mediated and non-immune factors, including inflammatory cytokines, ischemia, and aberrant signaling pathways.4,5 One of the key contributors to DM is the upregulation of type I interferons (IFNs), particularly IFNα, which is known to activate the JAK/STAT signaling pathway.6 Previous studies have shown that STAT1 is a key mediator of type I interferon-induced gene expression, which is highly upregulated in dermatomyositis-affected muscle tissues.7 In contrast, while STAT2 also participates in interferon signaling, its primary role is in complex formation with IRF9 rather than direct transcriptional regulation of pro-inflammatory and pro-atrophic genes.8 TYK2, as an upstream kinase, is more broadly involved in multiple cytokines signaling pathways beyond interferon signaling.9 The activation of STAT1, a crucial transcription factor in the type I IFN response, has been strongly implicated in the pathophysiology of DM.10,11 STAT1 mediates the expression of a range of interferon-stimulated genes (ISGs), which contribute to muscle inflammation, fibrosis, and impaired regeneration, thereby playing an essential role in DM-associated muscle pathology including perifascicular atrophy and muscle weakness.7,12,13
The ubiquitin-proteasome system (UPS) plays a critical role in regulating various cellular processes, including protein degradation, cell cycle control, and stress response.14 Ubiquitin E3 ligases are key components of the UPS, mediating the transfer of ubiquitin to specific substrate proteins, thereby targeting them for proteasomal degradation.15 In the context of DM, the role of ubiquitin-related processes in muscle damage and repair remains largely unknown. Recent evidence suggests that dysregulation of ubiquitination may contribute to muscle atrophy and impaired regeneration, as many key signaling pathways, including those involved in inflammation, differentiation, and apoptosis, are regulated by ubiquitination.16,17 However, the specific ubiquitin E3 ligases that may be involved in DM pathogenesis and their targets have not been systematically studied.
In this study, we aimed to investigate the role of ubiquitin E3 ligases in the muscle pathology of DM, focusing on the potential involvement of the IFNα/STAT1 signaling axis. By analyzing public datasets, we identified several ubiquitin E3 ligases that were significantly upregulated in DM muscle tissues, among which FBXO4 emerged as a candidate of particular interest. FBXO4 is an E3 ubiquitin ligase known to mediate the degradation of various substrates involved in cell cycle regulation and survival. We hypothesized that FBXO4 might play a role in DM muscle pathology by modulating key signaling pathways related to muscle cell differentiation and apoptosis. Our study aimed to elucidate the mechanisms through which FBXO4 contributes to DM pathogenesis, with a particular focus on its interaction with the IFNα/STAT1 pathway and its potential targets, such as β-catenin and MCL1. By understanding the role of FBXO4 in DM, we hoped to identify novel therapeutic targets to alleviate muscle damage and promote regeneration in affected patients.
Materials and Methods
Cell Culture and Treatments
Human immortalized skeletal muscle cell line LHCN-M2 was purchased from EVERCYTE and cultured in DMEM/M199 supplemented with 15% fetal bovine serum (FBS) and 1% penicillin-streptomycin. Mouse skeletal muscle cells (C2C12) and HEK-293T were purchased from the Cell Bank of the Chinese Academy of Sciences (CAS) and cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% FBS and 1% penicillin-streptomycin. Cells were maintained at 37 °C in a humidified atmosphere with 5% CO2. For IFNα treatments, cells were treated with 1000 U/mL IFNα for 24 hours to simulate inflammatory conditions. For Wnt/β-catenin signaling inhibition, cells were treated with 5 μM IWR-1-endo or DMSO as a control.
Gene Knockout and Overexpression
FBXO4 and STAT1 knockouts were performed using CRISPR/Cas9 technology. Guide RNAs (gRNAs) targeting FBXO4 and STAT1 were designed and cloned into lentiCRISPRv2 plasmid. Lentiviral particles were produced in HEK-293T cells and used to transduce LHCN-M2 cells. After selection with 2 μg/mL puromycin, knockout efficiency was confirmed by Western blotting and quantitative reverse transcription PCR (qRT-PCR).
For overexpression studies, STAT1 cDNA was cloned into the pCDH-CMV-MCS-EF1α-Puro vector. The constructs were transfected into HEK-293T cells using Lipofectamine 3000 (Thermo Fisher Scientific) following the manufacturer’s protocol.
RNA Sequencing and Data Analysis
RNA was extracted from control and FBXO4 knockout LHCN-M2 cells using the RNeasy Mini Kit (Qiagen). RNA sequencing analysis was performed by GENEWIZ life science. Briefly, for library construction, 1 µg of high-quality total RNA was used as input. mRNA was enriched using poly(A) selection with oligo(dT) magnetic beads for total RNA sequencing. The mRNA was then fragmented into short pieces (~200–300 bp) by incubation with divalent cations at elevated temperatures. First-strand cDNA synthesis was performed using reverse transcriptase (SuperScript III, Thermo Fisher) and random hexamer primers, followed by second-strand cDNA synthesis with DNA polymerase I and RNase H. Double-stranded cDNA was purified using AMPure XP beads (Beckman Coulter), followed by A-tailing and ligation of sequencing adapters (Illumina TruSeq RNA Library Prep Kit). After adapter ligation, libraries were size-selected (~350 bp) using AMPure XP beads, followed by PCR amplification (12–15 cycles) to enrich adapter-ligated fragments. The final library quality and concentration were assessed using the Agilent 2100 Bioanalyzer. And then the validated RNA-seq libraries were subjected to paired-end (PE) sequencing (150 bp reads) on an Illumina NovaSeq 6000 platform.
Differential gene expression analysis was conducted using DESeq2, with genes having a fold change > 2 and P value < 0.05 considered significantly differentially expressed. Gene ontology (GO) enrichment analysis was performed using the DAVID bioinformatics tool.
Quantitative Reverse Transcription PCR (qRT-PCR)
Total RNA was extracted from cells using the RNeasy Mini Kit (Qiagen). cDNA synthesis was performed using the iScript cDNA Synthesis Kit (Bio-Rad). qRT-PCR was conducted using SYBR Green Master Mix (Applied Biosystems) on a QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific). Relative gene expression was calculated using the ΔΔCt method, with 18S RNA as the reference gene. Primers used for qRT-PCR are listed in Table S1.
Western Blotting
Cells were lysed in SDS lysis buffer. Protein concentrations were determined using the BCA Protein Assay Kit (Thermo Fisher Scientific). Equal amounts of protein were separated by SDS-PAGE and transferred to Nitrocellulose membranes. Membranes were blocked with 5% non-fat milk and incubated with primary antibodies overnight at 4°C, followed by incubation with HRP-conjugated secondary antibodies. anti-ISG15, anti-FBXO4, anti-MYC and anti-β-catenin were purchased from Abcam. anti-CCND1, anti-MYOG, anti-MYOD, anti-SKP1 were purchased from Proteintech. Anti-Ub, anti-STAT1, anti-STAT2 and β-actin were purchased from CST. Membranes were visualized using an enhanced chemiluminescence (ECL) detection system (Bio-Rad).
Immunoprecipitation and Ubiquitination Assay
Cells were transfected with Flag-FBXO4 using Lipofectamine 2000 (Thermo Fisher Scientific). After 48 hours, cells were lysed in IP lysis buffer (Thermo Fisher Scientific) containing protease inhibitors. Lysates were incubated with anti-Flag antibody and protein G magnetic beads (Thermo Fisher Scientific) overnight at 4°C. Immunoprecipitates were washed and subjected to Western blotting to detect interacting proteins. For ubiquitination assays, cells were treated with 10 μM MG132 for 6 hours before lysis to inhibit proteasomal degradation, allowing detection of ubiquitinated proteins.
Luciferase Reporter Assay
The promoter region of FBXO4 (−3000 to 0 bp) was cloned into the pGL3-basic luciferase vector (Promega). Top-flash and Fop-flash luciferase vectors were purchased from Promega. HEK-293T cells were co-transfected with the luciferase construct and a Renilla luciferase plasmid as a control for transfection efficiency. After 24 hours, cells were treated with IFNα (1000 U/mL) for an additional 24 hours. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega), and results were normalized to Renilla luciferase activity.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assays were performed using the ChIP-IT Express Kit (Active Motif) according to the manufacturer’s instructions. LHCN-M2 cells were treated with IFNα (1000 U/mL) for 24 hours before crosslinking with 1% formaldehyde. Chromatin was sheared by sonication, and immunoprecipitation was performed using an anti-STAT1 antibody. qPCR was conducted to analyze the enrichment of STAT1 binding at the GAS sites in the FBXO4 promoter. Primers used for ChIP-qPCR are listed in Table S1.
Cell Morphology and Differentiation Assays
LHCN-M2 cells were cultured in differentiation medium (DMEM with 2% horse serum) for 5 days to induce differentiation. Cell morphology was observed under a phase-contrast microscope. Differentiation was assessed by immunofluorescence staining for myogenin (MYOG) and myosin heavy chain (MHC) using specific antibodies, followed by imaging with a fluorescence microscope (Nikon).
Cell Viability Assay
Cell viability was assessed using the Cell Counting Kit-8 (CCK-8, Dojindo) following the manufacturer’s instructions. Briefly, LHCN-M2 cells were seeded in 96-well plates and treated as indicated. CCK-8 solution was added to each well, and absorbance at 450 nm was measured after 2 hours using a microplate reader (BioTek).
Statistical Analysis
All experiments were performed at least three times independently. Data are presented as mean ± standard deviation (SD). Statistical analysis was conducted using GraphPad Prism 10.0 software. Differences between groups were analyzed using multiple t-test, with p < 0.05 considered statistically significant.
Results
Screening of Ubiquitin E3 Ligases in the Dermatomyositis Datasets
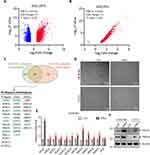
Although muscle damage in dermatomyositis (DM) is primarily caused by inflammation and ischemia due to immune system attacks on muscle fibers and blood vessels, little is known about the role of ubiquitin related biological process in DM-associated muscle damage. Therefore, this study aims to investigate whether ubiquitination-related enzymes, particularly ubiquitin E3 ligases, exhibit abnormal expression in the muscle tissues of DM patients by screening public databases. Differential gene expression analysis (fold change > 2, P-value < 0.01) was performed using two public datasets, GSE1197118 (Figure 1A) and GSE155119 (Figure 1B), comparing muscle tissues from DM patients to normal muscle tissues. Among the significantly upregulated genes, 53 and 87 ubiquitination-related proteins were identified in GSE11971 and GSE1551 datasets, respectively (Figure 1C), with 34 proteins found to be significantly elevated in both datasets (Figure 1C). Notably, several ubiquitin E3 ligases, including UBR2,20 DTX,21 WWP1,22 and deubiquitinating enzymes such as USP18,23 USP20,24,25 and USP22,26 which have been previously reported to play roles in muscle-related pathological or physiological processes, were identified. Thus, we hypothesize that the remaining unreported proteins may be involved in the pathogenesis of DM. To further explore this, human immortalized skeletal muscle cells (LHCN-M2) and a mouse myoblast cell line C2C12 were treated with the key inflammatory factor in DM, IFN-α, to simulate the effect of inflammatory factors on muscle cells during DM progression. We noticed that the cells exhibit a slender morphology, resembling the pathological state of perifascicular atrophy observed in muscle tissue (Figure 1D). Then qRT-PCR was employed to assess the expression of the selected, yet-to-be-reported, E3 ligases in DM. As expected, ISG15, a known STAT1 target gene,27 was significantly increased following IFNα treatment (Figure 1E), indicating the effectiveness of the IFNα treatment. All the detected E3 ligases showed varying degrees of upregulation after IFNα treatment (Figure 1E), with the most pronounced fold change observed in FBXO4 (Figure 1E), suggesting that FBXO4 might play a role in the pathogenesis of DM. In addition, the protein levels of FBXO4 were elevated in both LHCN-M2 and C2C12 cells, a mouse skeletal muscle cell line, following treatment with IFNα (Figure 1F). In conclusion, these findings indicate that the ubiquitin E3 ligase FBXO4 is significantly upregulated in the muscle tissues of DM patients, suggesting its involvement in the pathological progression of dermatomyositis.
Depletion of FBXO4 Promotes Myocyte Differentiation
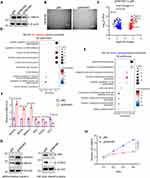
To investigate the role of FBXO4 in muscle cells, we knocked out FBXO4 in LHCN-M2 cells, as shown in Figure 2A, with gFBXO4#2 being more effective. We observed that, compared to the control, depletion of FBXO4 caused the morphology of LHCN-M2 cells to become larger and more elongated (Figure 2B), resembling a differentiated state. Subsequently, RNA sequencing revealed that the depletion of FBXO4 led to a significant increase in genes associated with muscle cell differentiation, such as MYOD1 and MYOG,28,29 while genes related to cell cycle, including E2F1, CCND1, and MYC, were significantly decreased (Figure 2C). Gene ontology enrichment analysis showed that the upregulated genes were significantly enriched in pathways related to muscle cell differentiation (Figure 2D), while the downregulated genes were enriched in cell proliferation and cell cycle pathways (Figure 2E). These results suggest that FBXO4 knockout induces differentiation in LHCN-M2 cells. qRT-PCR (Figure 2F) and Western blot (Figure 2G) analyses confirmed that the mRNA and protein levels of differentiation-related genes, such as MYOD1, MYOG, and MMP14, were significantly increased, while those related to cell proliferation, including MYC, CCND1, and E2F1, were markedly reduced. Furthermore, CCK8 assays validated that the depletion of FBXO4 LHCN-M2 cells was impaired following FBXO4 knockout (Figure 2H). Collectively, these findings demonstrate that the knockout of FBXO4 triggers terminal differentiation in muscle cells.
FBXO4 Ubiquitinates and Degrades β-Catenin in Muscle Cells
As a ubiquitin E3 ligase, we hypothesize that FBXO4 regulates muscle cell differentiation through the ubiquitination and degradation of its substrates. Using the online tool ubibrowser2.0, we analyzed the substrates of FBXO4. From the analysis results, it is known that the currently reported substrates of FBXO4 include CCND1,30 MCL1,31 ICAM,32 FXR1,33 and TERF1.32 Additionally, several potential target proteins were predicted, including those involved in the Wnt canonical signaling pathway, such as β-catenin, RELA, and p53 (Figure 3A). We noted that in the context of muscle injury, the Wnt/β-catenin signaling pathway is activated, promoting the proliferation and differentiation of muscle stem cells (satellite cells), thereby participating in muscle regeneration. Based on these clues, we speculate that in dermatomyositis, the elevated levels of FBXO4 might target and degrade β-catenin, thereby inhibiting the proliferation and differentiation of muscle stem cells, leading to sustained muscle damage. Subsequently, in LHCN-M2 cells, immunoprecipitation experiments following the exogenous expression of Flag-FBXO4 confirmed that FBXO4 indeed interacts with β-catenin, MCL1 and SKP1 were used as positive control here (Figure 3B). Furthermore, depletion of FBXO4 elevated the protein level of β-catenin (Figure 3C). These results indicate that FBXO4 might regulate the degradation of β-catenin through ubiquitination. We then examined the ubiquitinated level of β-catenin in FBXO4 knockout and control cells. As shown in Figure 3D, knockout of FBXO4 reduces the ubiquitinated β-catenin compared to control. These results verified that β-catenin is a direct target of FBOX4. We then asked whether FBXO4 impaired Wnt/β-catenin signaling in LHCN-M2 cells. Luciferase reporter assays revealed that depletion of FBXO4 enhances the Top-flash luciferase activity in LHCN-M2 cells (Figure 3E), indicates that depletion of FBXO4 promotes Wnt/β-catenin signaling. Altogether, these findings suggested that FBXO4 binds to β-catenin and promotes its degradation by ubiquitination.
Depletion of FBXO4 Promotes Muscle Cell Differentiation Through Wnt/β-Catenin Signaling
We investigated whether FBXO4 inhibits muscle cell differentiation via the Wnt/β-catenin pathway. FBXO4 knockout and control LHCN-M2 cells were treated with the Wnt signaling inhibitor IWR-1-endo34 or DMSO. Compared to DMSO, FBXO4 knockout cells treated with IWR-1-endo exhibited reduced cell size (Figure 4A), suggesting that IWR-1-endo restored the differentiation state induced by FBXO4 loss. Additionally, mRNA (Figure 4B) and protein levels (Figure 4C) of differentiation markers MYOG, MYOD and MMP14 were significantly rescued by IWR-1-endo in FBXO4 knockout cells. These findings indicate that FBXO4 depletion promotes muscle cell differentiation through activation of the Wnt/β-catenin pathway.
We further considered that FBXO4 targets MCL1, an anti-apoptotic member of the BCL2 family,35 and hypothesized that in differentiation medium (DM), upregulated FBXO4 may promote MCL1 degradation, leading to muscle cell apoptosis. To test this, we assessed apoptosis in FBXO4 knockout and control LHCN-M2 cells treated with IFNα, and found that FBXO4 depletion inhibited cell death under IFNα treatment. These findings indicate that in DM, highly expressed FBXO4 promotes muscle cell apoptosis while inhibiting muscle cell differentiation.
In the other hand, we considered that FBXO4 targets MCL1, an anti-apoptotic member of the BCL2 family, and hypothesized that in DM, upregulated FBXO4 may promote MCL1 degradation, leading to muscle cell apoptosis. To clarify this, we evaluated apoptosis in both FBXO4 knockout and control LHCN-M2 cells after treatment with IFNα and found that FBXO4 depletion inhibited cell death under IFNα treatment (Figure 5). These findings indicate that in DM, highly expressed FBXO4 promotes muscle cell apoptosis while inhibiting muscle cell differentiation.
FBXO4 Is Induced by IFNα/STAT1 Signaling in DM
We investigated how FBXO4 is upregulated in dermatomyositis (DM). Previous results (Figure 1D) suggested that FBXO4 could be induced by IFNα, leading us to hypothesize that FBXO4 might be a target of the IFNα/JAK/STAT1 pathway. We treated LHCN-M2 cells with Fludarabine, a STAT1 signaling inhibitor,36 and observed that it significantly reversed the increase in FBXO4 levels under IFNα treatment (Figure 6A), supporting that FBXO4 is downstream of STAT1 signaling. We then knocked out STAT1 in LHCN-M2 cells, treated them with IFNα, and found that STAT1 depletion significantly reduced both the mRNA (Figure 6B) and protein levels (Figure 6C) of FBXO4 under IFNα treatment. Moreover, knockout of TYK2—the upstream kinase of STAT1 and STAT2—significantly reduced FBXO4 expression, whereas STAT2 knockout did not (Figure S1A). In addition, STAT1 knockout resulted in a cell morphology similar to that observed with FBXO4 knockout (Figure S1B). Together, these findings suggest that FBXO4 is a downstream target of STAT1.
Additionally, promoter analysis of FBXO4 using the TFAP online tool identified six GAS (Gamma-Activated Sequence) sites, which are STAT1 binding sites (Figure 6D), suggesting FBXO4 as a potential direct target of STAT1. We cloned the promoter sequence (−3000 to 0) of FBXO4 into the pGL3-basic vector, overexpressed STAT1 in HEK-293T cells (Figure 6D), and conducted dual-luciferase assays under IFNα treatment. As shown in Figure 6E, STAT1 overexpression significantly increased FBXO4 promoter activity, with pSTAT1-TA-Luc serving as a positive control (Figure 6E).
Further, STAT1 ChIP-PCR was performed in HEK-293T cells after IFNα treatment. Western blot confirmed the ChIP assays (Figure 6F), and ChIP-qPCR demonstrated that STAT1 binds to the GAS1-3 and GAS5-6 sites of the FBXO4 promoter (Figure 6D and G). ISG15 was included as a positive control. In conclusion, these findings reveal that FBXO4 is induced via the IFNα-JAK-STAT1 pathway in DM and is a direct target of STAT1.
Discussion
In this study, to investigate the role of ubiquitin pathway–related proteins in muscle damage associated with DM, we screened publicly available datasets to identify ubiquitin E3 ligases and Deubiquitinases that are differentially expressed in DM compared to normal muscle tissue. A total of 34 ubiquitin-related proteins were identified, including UBR2,20 USP18,23 and USP20,25 which have been previously reported to participate in muscle-related physiological or pathological processes, supporting the reliability of our screening strategy. Among these, we focused on the subclass of ubiquitin E3 ligases and identified FBXO4 as the most significantly upregulated gene at the mRNA level in an IFNα-induced cell model. Subsequent experiments demonstrated that elevated FBXO4 promoting β-catenin degradation, thereby inhibiting the canonical Wnt signaling pathway and ultimately suppresses muscle cell differentiation.
The Wnt/β-catenin signaling pathway is essential for normal organ development.37 In skeletal muscle regeneration, activated Wnt/β-catenin signaling by promoting muscle-specific gene expression, supporting satellite cell self-renewal, and regulating the transition from proliferation to myogenic differentiation.38,39 In the current study, we identified that β-catenin is a substrate of FBXO4. Persistent IFNα/JAK/STAT1 signaling inducing the expression of FBXO4 and thereby promotes the degradation of β-catenin and thereby inhibits muscle cell differentiation.
In DM, chronic inflammation, persistent type I interferon signaling, and associated pathological changes disrupt the normal regeneration and differentiation of myoblasts.40 High expression of FBXO4 inhibits muscle cell differentiation, thereby impairing muscle repair and regeneration and ultimately promoting the progression of DM.
However, there remain some concerns that should be further investigated in the future:1) all the experiments were done in cell models, with no animal and patient samples to assess the expression of FBXO4 and its association with Wnt/β-catenin signaling, myoblast differentiation and perifascicular atrophy in DM; 2) the detailed mechanism of FBXO4 degrades β-catenin, such as the binding site mapping; 3) Following the resolution of the above issues, we will screen for small molecules that disrupt the FBXO4–β-catenin interaction, aiming to identify novel therapeutics for relieving muscle weakness in dermatomyositis patients.
Conclusion
Our study highlights the significance of FBXO4 as a key regulator of muscle cell fate in DM, linking inflammatory signaling with ubiquitin-mediated degradation pathways. The upregulation of FBXO4 in response to IFNα suggests that inflammatory cytokines may drive muscle pathology in DM by modulating ubiquitin E3 ligase activity. Targeting FBXO4 or its downstream substrates, such as β-catenin and MCL1, could provide novel therapeutic strategies to promote muscle regeneration and prevent apoptosis in DM. Future studies should explore the potential of FBXO4 inhibitors or strategies to stabilize β-catenin and MCL1 as possible treatments for DM, aiming to restore the balance between muscle regeneration and apoptosis in affected patients.
Data Sharing Statement
The relevant data and materials are available from the first author Liguo Yin (E-mail: [email protected]) upon reasonable request.
Ethics Statement
This study was approved by the Ethics Committee of the Shandong Provincial Hospital Affiliated to Shandong First Medical University.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This research was funded by the Natural Science Foundation of Shandong Province (Youth fund project, Grant No. ZR2021QH184).
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Suárez-Calvet X, Gallardo E, Pinal-Fernandez I, et al. RIG-I expression in perifascicular myofibers is a reliable biomarker of dermatomyositis. Arthritis Res Ther. 2017;19(1):174. doi:10.1186/s13075-017-1383-0
2. DeWane ME, Waldman R, Lu J. Dermatomyositis: clinical features and pathogenesis. J Am Acad Dermatol. 2020;82(2):267–281. doi:10.1016/j.jaad.2019.06.1309
3. Waldman R, DeWane ME, Lu J. Dermatomyositis: diagnosis and treatment. J Am Acad Dermatol. 2020;82(2):283–296. doi:10.1016/j.jaad.2019.05.105
4. Lundberg IE, Fujimoto M, Vencovsky J, et al. Idiopathic inflammatory myopathies. Nat Rev Dis Primers. 2021;7(1):86. doi:10.1038/s41572-021-00321-x
5. Kim H. Updates on interferon in juvenile dermatomyositis: pathogenesis and therapy. Curr Opin Rheumatol. 2021;33(5):371–377. doi:10.1097/bor.0000000000000816
6. Kao L, Chung L, Fiorentino DF. Pathogenesis of dermatomyositis: role of cytokines and interferon. Curr Rheumatol Rep. 2011;13(3):225–232. doi:10.1007/s11926-011-0166-x
7. Zhang SH, Zhao Y, Xie QB, Jiang Y, Wu YK, Yan B. Aberrant activation of the type I interferon system may contribute to the pathogenesis of anti-melanoma differentiation-associated gene 5 dermatomyositis. Br J Dermatol. 2019;180(5):1090–1098. doi:10.1111/bjd.16917
8. Duodu P, Sosa G, Canar J, Chhugani O, Gamero AM. Exposing the two contrasting faces of STAT2 in inflammation. J Interferon Cytokine Res. 2022;42(9):467–481. doi:10.1089/jir.2022.0117
9. Rusiñol L, Puig L. Tyk2 targeting in immune-mediated inflammatory diseases. Int J Mol Sci. 2023;24(4):3391. doi:10.3390/ijms24043391
10. Alves P, Bashir MM, Wysocka M, Zeidi M, Feng R, Werth VP. Quinacrine suppresses tumor necrosis factor-α and IFN-α in dermatomyositis and cutaneous lupus erythematosus. J Investig Dermatol Symp Proc. 2017;18(2):S57–s63. doi:10.1016/j.jisp.2016.11.001
11. Moneta GM, Pires Marafon D, Marasco E, et al. Muscle expression of type I and type II interferons is increased in juvenile dermatomyositis and related to clinical and histologic features. Arthritis Rheumatol. 2019;71(6):1011–1021. doi:10.1002/art.40800
12. Wilkinson MGL, Moulding D, McDonnell TCR, et al. Role of CD14+ monocyte-derived oxidised mitochondrial DNA in the inflammatory interferon type 1 signature in juvenile dermatomyositis. Ann Rheum Dis. 2023;82(5):658–669. doi:10.1136/ard-2022-223469
13. Tanboon J, Inoue M, Saito Y, et al. Dermatomyositis: muscle pathology according to antibody subtypes. Neurology. 2022;98(7):e739–e749. doi:10.1212/wnl.0000000000013176
14. Pohl C, Dikic I. Cellular quality control by the ubiquitin-proteasome system and autophagy. Science. 2019;366(6467):818–822. doi:10.1126/science.aax3769
15. Zheng N, Shabek N. Ubiquitin ligases: structure, function, and regulation. Annu Rev Biochem. 2017;86:129–157. doi:10.1146/annurev-biochem-060815-014922
16. Liu Q, Xu WG, Luo Y, et al. Cigarette smoke-induced skeletal muscle atrophy is associated with up-regulation of USP-19 via p38 and ERK MAPKs. J Cell Biochem. 2011;112(9):2307–2316. doi:10.1002/jcb.23151
17. Liu X, Zheng T, Zhang Y, et al. Endothelial Dickkopf-1 promotes smooth muscle cell-derived foam cell formation via usp53-mediated deubiquitination of SR-A during atherosclerosis. Int J Biol Sci. 2024;20(8):2943–2964. doi:10.7150/ijbs.91957
18. Chen YW, Shi R, Geraci N, Shrestha S, Gordish-Dressman H, Pachman LM. Duration of chronic inflammation alters gene expression in muscle from untreated girls with juvenile dermatomyositis. BMC Immunol. 2008;9:43. doi:10.1186/1471-2172-9-43
19. Greenberg SA, Pinkus JL, Pinkus GS, et al. Interferon-alpha/beta-mediated innate immune mechanisms in dermatomyositis. Ann Neurol. 2005;57(5):664–678. doi:10.1002/ana.20464
20. Gao S, Zhang G, Zhang Z, et al. UBR2 targets myosin heavy chain IIb and IIx for degradation: molecular mechanism essential for cancer-induced muscle wasting. Proc Natl Acad Sci U S A. 2022;119(43):e2200215119. doi:10.1073/pnas.2200215119
21. Wang Y, Yang BP, Chi YG, Liu LB, Lei L. Effect of Deltex-1 on proliferation and differentiation of bone marrow mesenchymal stem cells into smooth muscle cells. Eur Rev Med Pharmacol Sci. 2018;22(12):3627–3634. doi:10.26355/eurrev_201806_15239
22. Hirata Y, Nomura K, Senga Y, et al. Hyperglycemia induces skeletal muscle atrophy via a WWP1/KLF15 axis. JCI Insight. 2019;4(4). doi:10.1172/jci.insight.124952
23. Olie CS, Pinto-Fernández A, Damianou A, et al. USP18 is an essential regulator of muscle cell differentiation and maturation. Cell Death Dis. 2023;14(3):231. doi:10.1038/s41419-023-05725-z
24. Zhang L, Wu JH, Jean-Charles PY, et al. Phosphorylation of USP20 on Ser334 by IRAK1 promotes IL-1β-evoked signaling in vascular smooth muscle cells and vascular inflammation. J Biol Chem. 2023;299(7):104911. doi:10.1016/j.jbc.2023.104911
25. Jean-Charles PY, Wu JH, Zhang L, et al. USP20 (Ubiquitin-Specific Protease 20) inhibits TNF (Tumor Necrosis Factor)-triggered smooth muscle cell inflammation and attenuates atherosclerosis. Arterioscler Thromb Vasc Biol. 2018;38(10):2295–2305. doi:10.1161/atvbaha.118.311071
26. Jiang YN, Yang SX, Guan X, et al. Loss of USP22 alleviates cardiac hypertrophy induced by pressure overload through HiF1-α-TAK1 signaling pathway. Biochim Biophys Acta Mol Basis Dis. 2023;1869(8):166813. doi:10.1016/j.bbadis.2023.166813
27. Forys JT, Kuzmicki CE, Saporita AJ, Winkeler CL, Maggi LB Jr, Weber JD. ARF and p53 coordinate tumor suppression of an oncogenic IFN-β-STAT1-ISG15 signaling axis. Cell Rep. 2014;7(2):514–526. doi:10.1016/j.celrep.2014.03.026
28. Liu Z, Zhang X, Lei H, et al. CASZ1 induces skeletal muscle and rhabdomyosarcoma differentiation through a feed-forward loop with MYOD and MYOG. Nat Commun. 2020;11(1):911. doi:10.1038/s41467-020-14684-4
29. Lei S, Li C, She Y, Zhou S, Shi H, Chen R. Roles of super enhancers and enhancer RNAs in skeletal muscle development and disease. Cell Cycle. 2023;22(5):495–505. doi:10.1080/15384101.2022.2129240
30. Kanie T, Onoyama I, Matsumoto A, et al. Genetic reevaluation of the role of F-box proteins in cyclin D1 degradation. Mol Cell Biol. 2012;32(3):590–605. doi:10.1128/mcb.06570-11
31. Feng C, Yang F, Wang J. FBXO4 inhibits lung cancer cell survival by targeting Mcl-1 for degradation. Cancer Gene Ther. 2017;24(8):342–347. doi:10.1038/cgt.2017.24
32. Kang JH, Choi MY, Cui YH, et al. Regulation of FBXO4-mediated ICAM-1 protein stability in metastatic breast cancer. Oncotarget. 2017;8(47):83100–83113. doi:10.18632/oncotarget.20912
33. Qie S, Majumder M, Mackiewicz K, et al. Fbxo4-mediated degradation of Fxr1 suppresses tumorigenesis in head and neck squamous cell carcinoma. Nat Commun. 2017;8(1):1534. doi:10.1038/s41467-017-01199-8
34. Nair S, Davis A, Campagne O, Schuetz JD, Stewart CF. Development and validation of a sensitive and specific LC-MS/MS method for IWR-1-endo, a wnt signaling inhibitor: application to a cerebral microdialysis study. Molecules. 2022;27(17):5448. doi:10.3390/molecules27175448
35. Li S, Wang J, Hu G, et al. SUMOylation of MCL1 protein enhances its stability by regulating the ubiquitin-proteasome pathway. Cell Signal. 2020;73:109686. doi:10.1016/j.cellsig.2020.109686
36. Martinez-Lostao L, Briones J, Forné I, et al. Role of the STAT1 pathway in apoptosis induced by fludarabine and JAK kinase inhibitors in B-cell chronic lymphocytic leukemia. Leuk Lymphoma. 2005;46(3):435–442. doi:10.1080/10428190400018398
37. Rim EY, Clevers H, Nusse R. The wnt pathway: from signaling mechanisms to synthetic modulators. Annu Rev Biochem. 2022;91:571–598. doi:10.1146/annurev-biochem-040320-103615
38. Petropoulos H, Skerjanc IS. Beta-catenin is essential and sufficient for skeletal myogenesis in P19 cells. J Biol Chem. 2002;277(18):15393–15399. doi:10.1074/jbc.M112141200
39. Tanaka S, Terada K, Nohno T. Canonical wnt signaling is involved in switching from cell proliferation to myogenic differentiation of mouse myoblast cells. J Mol Signal. 2011;6:12. doi:10.1186/1750-2187-6-12
40. André LM, Ausems CRM, Wansink DG, Wieringa B. Abnormalities in skeletal muscle myogenesis, growth, and regeneration in myotonic dystrophy. Front Neurol. 2018;9:368. doi:10.3389/fneur.2018.00368
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.