Back to Journals » Journal of Inflammation Research » Volume 18
Immune Microenvironment Characterization and Machine Learning-Guided Identification of Diagnostic Biomarkers for Ulcerative Colitis
Authors Zheng Q, Wang L, Zhang Y , Peng J, Hou J, Wang H, Ma Y, Tang P, Li Y, Li H, Chen Y, Li J, Chen Y
Received 26 March 2025
Accepted for publication 27 June 2025
Published 9 July 2025 Volume 2025:18 Pages 8977—8992
DOI https://doi.org/10.2147/JIR.S526325
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Nadia Andrea Andreani
Qingqing Zheng,1,2,* Li Wang,1,2,* Yu Zhang,3,* Jun Peng,4,* Jianhong Hou,4,* Hui Wang,1,2 Yazhe Ma,5 Peiren Tang,1,2 Ying Li,1,2 Huan Li,1,2 Yun Chen,4 Jie Li,6 Yang Chen1,2,7
1Department of Pathology, The First People’s Hospital of Yunnan Province, Kunming, Yunnan, 650032, People’s Republic of China; 2Department of Pathology, The Affiliated Hospital of Kunming University of Science and Technology, Kunming, Yunnan, 650032, People’s Republic of China; 3Department of Gastroenterology, The First People’s Hospital of Yunnan Province, Kunming, Yunnan, 650032, People’s Republic of China; 4Department of Surgery, The First People’s Hospital of Yunnan Province, Kunming, Yunnan, 650032, People’s Republic of China; 5Yunnan Arrhythmia Research Center, Division of Cardiology, The First People’s Hospital of Yunnan Province, Kunming, Yunnan, 650032, People’s Republic of China; 6Academy of Biomedical Engineering, Kunming Medical University, Kunming, Yunnan, 650500, People’s Republic of China; 7Yunnan Provincial Laboratory of Clinical Virology, The First People’s Hospital of Yunnan Province, Kunming, Yunnan, 650032, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yang Chen, The First People’s Hospital of Yunnan Province, Kunming, 650032, People’s Republic of China, Email [email protected] Jie Li, Academy of Biomedical Engineering, Kunming Medical University, Kunming, Yunnan, 650500, People’s Republic of China, Email [email protected]
Background: Ulcerative colitis (UC) is a chronic inflammatory bowel disease hallmarked by dysregulated immune responses. Current treatments often show limited efficacy, highlighting the need for novel diagnostic and therapeutic approaches.
Methods: RNA-Seq data from 495 UC patients and 320 controls (training dataset) and 389 UC patients and 209 controls (testing dataset) were analyzed. Immune cell infiltration was assessed via the ImmuCellAI algorithm, while differential expression analysis and WGCNA were performed to identify key immune-related genes. Moreover, machine learning models, including Random Forest and Best Subset Selection, were used to construct and validate an optimal diagnostic framework. Lastly, the findings were further corroborated using immunohistochemistry conducted on tissue samples from UC patients and controls.
Results: Thirteen immune cell types, including B cells, macrophages, and naive CD4+ T cells, were identified as significantly altered in UC. Likewise, cytokines such as IL-10, TGF-β, RORγ, and IL-21 exhibited abnormal expression patterns in UC tissues. WGCNA identified three immune cell-associated gene modules, among which the MEblue, MEturquoise, and MEgrey modules were highly correlated with aberrant immune cells. Additionally, machine learning models identified 99 candidate genes, from which an optimal diagnostic model comprising eight crucial genes (GATA2, IL8, LAT, NOLC1, SMARCA5, SMC3, STX10, ZMIZ1) was constructed, achieving an AUC of 0.964 in the training dataset, 0.926 in the internal test dataset, and 0.884 in the independent test dataset. Functional enrichment analysis revealed associations with inflammatory and immune-regulatory pathways, highlighting their biological relevance. Moreover, the identified eight genes hold translational potential for clinical diagnostics and may serve as a foundation for future precision-targeted therapies in UC.
Conclusion: This study highlights alterations in the immune microenvironment in UC and presents an accurate eight-gene diagnostic model, offering the potential for early detection and novel therapeutic targets.
Plain Language Summary: What is Already Known: Previous studies have established that UC involves immune dysregulation, including impaired intestinal barrier function, immune cell infiltration, and alterations in cytokine expression. Conventional treatments primarily focus on anti-inflammatory strategies but are often limited by relapses and a lack of durability.
What is new here: This study identifies a distinct pattern of immune cell dysregulation in UC patients, involving abnormalities in macrophages, neutrophils, and T-cell subsets. It employs machine learning algorithms to construct diagnostic models, including an optimal 8-gene model (GATA2, IL8, LAT, NOLC1, SMARCA5, SMC3, STX10, ZMIZ1), which demonstrates high predictive performance (AUC of 0.964 in training datasets and 0.884 in testing datasets). Functional validation confirmed the abnormal expression of cytokines associated with the immune imbalance in UC.
How can this study help patient care: This research benefits clinicians, researchers, and pharmaceutical developers by providing insights into the immunopathogenesis of UC. It highlights potential diagnostic biomarkers and therapeutic targets, aiding in the development of precision medicine approaches for UC management.
Keywords: ulcerative colitis, immune microenvironment, machine learning, diagnostic biomarkers
Graphical Abstract:

Introduction
Ulcerative colitis (UC), a form of inflammatory bowel disease (IBD), mainly affects the mucosal and submucosal layers of the rectum and colon. Its primary symptoms include abdominal discomfort, diarrhea, and stools containing mucus, pus, or blood. The disease is characterized by alternating episodes of remissions and relapses and affects the rectum and sigmoid colon, predominantly in young adults aged 20–40 years old, and is a prevalent disease of the digestive system.1,2 Its pathogenesis involves a complex set of etiological and pathophysiological factors, including environmental factors, genetic variation, intestinal microbial imbalance, intestinal inflammation, immune dysregulation, etc. Despite its intricate pathogenesis, the exact reason underlying the high prevalence of UC remains elusive. Among its contributing factors, immune dysregulation is considered a dominant driver of the pathogenesis of UC.3 The immunopathogenesis of UC can be summarized as follows: impaired intestinal barrier function, loss of immune tolerance to intestinal antigens, marked infiltration of adaptive immune cells into the intestinal lamina propria, alteration of the immune response, activation of inflammatory pathways, up-regulation of the expression of proinflammatory cytokines, and an anti-inflammatory/proinflammatory imbalance.4 Immune cells constitute an integral component of the body’s immune system, which not only defends against foreign germs but also removes senescent or dysregulated cells. In UC patients, the immune system is generally over-activated by various factors, with the influx of immune cells into the intestines culminating in the destruction of intestinal tissues.5 In recent years, the immune microenvironment in UC has also garnered extensive research attention. It is a complex system containing various cell types interacting with each other, thereby forming an orderly spatial relationship.6,7
Dysregulation of the immune system influenced by genetic and environmental factors and gut microbiota is closely associated with the progression of UC.8 The intestinal immune microenvironment consists of intestinal epithelial cells, macrophages, dendritic cells (DCs), regulatory T cells (Tregs), and inflammatory T cells, which collectively work to maintain immune homeostasis.9 Targeting the immune microenvironment may provide therapeutic benefits for UC. For example, macrophages (MΦ) play a key role in the progression of UC.10 They have traditionally been categorized into M1Φ with pro-inflammatory/anti-microbial activity and M2Φ with anti-inflammatory/tissue repair activity.11 An increase in the proportion of M1Φ at pathological sites of colitis has been correlated with disease progression.12 In addition to the inflammatory immune response exacerbated by colitis, hyperproliferation of fibroblasts and myofibroblasts contributes to extracellular matrix (ECM) deposition. While conventional treatment is largely based on anti-inflammatory interventions (eg, 5-aminosalicylic acid, corticosteroids, and immunosuppressants) to alleviate symptoms, their clinical use is limited by the lack of long-lasting efficacy and relapse.13 Although these immunological insights have advanced, UC diagnosis in clinical practice still relies heavily on endoscopy and non-specific biomarkers such as CRP and fecal calprotectin, which lack disease specificity and fail to reflect immune heterogeneity. Moreover, clinicians face difficulties in predicting disease trajectory or therapeutic response based on current diagnostic tools. To address these challenges, recent advances have highlighted the potential of reactive oxygen species (ROS)-responsive nanocarriers and biomimetic delivery systems to achieve localized, on-demand drug release within the inflamed intestinal microenvironment. These systems leverage the oxidative stress characteristic of ulcerative colitis to selectively activate drug release, thereby enhancing therapeutic precision and minimizing off-target effects. These emerging strategies hold promise for integrating diagnostic markers with therapeutic modalities to enable precision medicine in UC management.14
In this study, we aimed to comprehensively characterize the UC immune microenvironment using RNA-seq–based immune cell deconvolution and weighted gene co-expression network analysis (WGCNA). We further employed machine learning algorithms to construct a diagnostic model and identify key genes with potential relevance to immune regulation and therapeutic response (Figure 1). Overall, this study lays a foundation for expanding our understanding of immune microenvironmental changes in UC and offers potential biomarkers for its diagnosis and treatment.
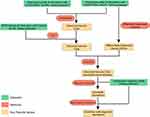 |
Figure 1 Workflow of the study. |
Materials and Methods
Patients and Control Subjects
The RNA-Seq data of 495 UC patients and 320 healthy controls was retrieved from the GEO database under accession number GSE177044 and used as the training dataset. Another independent testing dataset containing data on 389 UC patients and 209 healthy controls was downloaded under accession number GSE186507 (Testing dataset 1). All FPKM expression values were normalized using a log2 transformation.
An additional dataset was collected from The First People’s Hospital of Yunnan Province for experimental validation. FFPE samples from 24 patients with UC and 21 healthy controls were collected between January 2022 and December 2023. To minimize the impact of confounding variables, patients with recent immunosuppressive treatment or co-existing inflammatory conditions were excluded. These samples were used for subsequent HE and IHC verification (Testing dataset 2). The FFPE samples’ ethical approval for this study was granted by the Ethics Committee of the First People’s Hospital of Yunnan Province (KHLL2024-KY199). Otherwise, since this study did not require any intervention or experiment related to patients, no informed consent was required after review by the Ethics Committee of the First People’s Hospital of Yunnan Province.
Hematoxylin-Eosin (H&E) Staining and Immunohistochemistry (IHC)
Sigmoid colon tissues from 24 patients with UC and 21 healthy controls were selected according to inclusion criteria that excluded recent anti-inflammatory or immunomodulatory treatments. All patients had no history of malignancy or other autoimmune disorders. The samples were sequentially fixed with 4% paraformaldehyde, paraffin-embedded, sectioned into 4-mm-thick sections, deparaffinized, rehydrated, and stained with hematoxylin and eosin (H&E).
Next, they were incubated with primary antibodies (FOXP3 Rabbit pAb, Zenbio, catalog number 251365, 1:50 dilution; ROR gamma T Rabbit mAb, Zenbio, catalog number R50198, 1:100 dilution; TGF beta 1 Rabbit pAb, Zenbio, catalog number 346599, 1:50 dilution; IL-10 Rabbit pAb, Zenbio, catalog number 502171, 1:50 dilution; IL-21 Antibody, Affinity, catalog number DF4818, 1:100 dilution) overnight at 4°C. Subsequently, the sections were washed three times with PBS and incubated with a secondary antibody (MaxvisionTM3 HRP-Polymer IHC Kit, MXB, catalog number KIT-5220) for 30 minutes at room temperature. The staining signal was visualized using a chromogenic agent and counterstained with hematoxylin. After dehydration, the sections were blocked using a permanent mounting medium and allowed to dry before observation under a microscope.
Then, microscopic counting and optical density scoring (OD) were performed to assess the expression levels of the aforementioned antibodies in UC patients and normal controls. Regarding microscopic counting, positive cells were quantified by manually counting the number of stained cells in five randomly selected high-power fields (HPFs, 40× magnification) under a light microscope. The average count per HPF was recorded as the IHC Microscopic Count. For the OD scoring, the intensity of IHC staining was semi-quantitatively evaluated using ImageJ software. Images of stained sections were captured under identical conditions, and the mean optical density (integrated optical density/area) was calculated for each sample. The OD score reflects staining intensity, providing a quantitative measure of protein expression. Both microscopic count and OD score were independently evaluated by two blinded observers to ensure consistency and minimize subjective bias. Discrepancies were resolved by a third observer. Statistical analysis was conducted to compare expression levels between groups.
Immune Microenvironment Analysis
ImmuCellAI, a tool that estimates the abundance of 24 immune cells, was utilized to determine the distribution of immune cells in UC patients. In the current study, the abundance of these 24 immune cells in UC patients were calculated and the compared with healthy controls.
Differential Expression Analysis
The limma R package was utilized to examine gene expression variations in UC patients. Genes were identified as differentially expressed when adjusted P_value < 0.05 and absolute log2fold_change ≥ 0.585.
WGCNA
Co-expression modules were selected using the WGCNA R package to investigate candidate genes exhibiting correlated patterns in expression levels, as well as their relationship with abnormal immune cell types in UC patients. Hierarchical clustering was employed to classify genes based on similarities in expression profiles. The resulting gene clusters were associated with dysregulated immune cells in patients, and key genes within these clusters were selected for further analysis.
Machine Learning
The Random Forest algorithm was applied to identify candidate genes within immune-cell-associated co-expression modules in UC patients. Initially, the training dataset was randomly split into a training set and a self-test set in a 7:3 ratio and a Random Forest (RF) model was developed for feature selection. The RF model combined multiple decision tree classifiers trained on random subsets of features from the training dataset and was implemented using the randomForest R package, with the number of trees set to 1000. The random seed was set to 123 to ensure reproducibility. To ensure robustness, 10-fold cross-validation was conducted on the dataset for internal validation. Then, the SVM model was constructed based on crucial genes selected through RF (which have lowest error.cv), and model performance was examined in both the training and testing datasets.
Subsequently, a Best Subset Selection regression model was employed. This method evaluates all possible combinations of predictor variables and selects the optimal model based on specific criteria such as adjusted R2, or BIC. Higher adjusted R2 and lower BIC values indicate a superior model. The Leaps R package was used to perform the Best Subset Selection, ultimately constructing the most effective diagnostic model for UC.
This study was conducted in accordance with the TRIPOD (Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis) guidelines to ensure rigorous and reproducible development and validation of the diagnostic model.
Function Enrichment Analysis
All differentially expressed genes (DEGs) were identified and subjected to functional enrichment analysis using the clusterprofiler R package. The analysis focused on KEGG pathways and Hallmark gene sets, with enriched functions selected based on a false discovery rate (FDR) less than 0.05.
Then, GSEA analysis was performed on crucial genes, which were filtered by the Best Subset Selection regression model using the clusterprofiler R package.
Statistical analyses were performed using R software (version 4.3.2). Group comparisons were analyzed with a t-test, and adjusted P_value less than 0.05 was considered statistically significant.
Results
Immune Microenvironment in UC Patients
To begin, the immune microenvironment of 495 UC patients and 320 healthy controls was analyzed based on the expression data (Training dataset). The results revealed that the abundance of B cells, CD8_naive cells, exhausted T cells, macrophages, and neutrophils was significantly higher in UC patients, whereas that of, Gamma_delta T, CD8_T, Tfh (T follicular helper cell), CD4_naive, iTreg (induced regulatory T cell), Th1, Th2, NKT, cytotoxic, MAIT, Central_memory T, CD4_T, Tr1 (type 1 regulatory T cell), Th17, and Effector_memory T cells were significantly lower (Figure 2A). Thereafter, these results were validated in another dataset consisting of 389 UC patients and 209 healthy controls (Testing dataset 1). Consistent with the results of the training dataset, the abundance of B cells, CD4_naive cells, CD4_T, CD8_naive cells, cytotoxic cells, effector_memory T cells, gamma_delta T, iTreg, Macrophage, Th2, Th17, and Tr1 was abnormal in UC patients (Figure 2B). Additionally, dysregulated immune cells in both the training and test datasets were screened, yielding 13 immune cells that were abnormal in UC (Supplemental Figure 1).
Furthermore, the abnormal immune cell dysregulation was analyzed in UC patients. Notably, CD4+ naive T cells can differentiate into various subtypes in response to cytokines. They exert immune-activating or immunosuppressive effects by binding to MHC II-peptide. Under the action of TGF-β, CD4+ naive T cells are transformed into iTreg and synthesize Tr1. Both iTreg and Tr1 are representative immunosuppressive cells that can exert immunosuppressive effects under the action of IL-10, TGF-β and FOXP. Besides, elevated macrophage infiltration and reduced gamma_delta T cells in UC patients can also be activated under the action of IL-10, TGF-β and FOXP3 and play an immunosuppressive and inflammatory role in response to IL-10. CD4+ naive T cells can also differentiate into Th17 and Tfh in response to TGF-β and IL-21, and both are involved in autoimmunity and germinal center formation in response to IL-21 and RORγ. Among them, autoimmunity was also associated with neutrophils, which were abnormally increase in UC in this study. Likewise, CD4+ naive cells T producing Th2 was abnormally decrease in UC patients in this study and participated in immune responses mediated by IL-10. Similarly, B cells were abnormal in UC patients and were associated with immune responses (Figure 3A).
Finally, the expression levels of key cytokines in the immune cell flows described earlier were validated in 24 UC patients and 21 healthy controls (Testing dataset 2). Specifically, the expression of RORγ, FOXP3, TGF-β, IL-10, and IL-21 was examined (Figure 3B). Next, the expression of these markers between UC patients and healthy controls was compared via microscopic counting and optical density scoring (OD). The results demonstrated that RORγ, FOXP3, TGF-β, IL-10 and IL-21 were abnormally expressed in UC patients using both methods, suggesting the presence of an abnormal immune microenvironment in UC patients, which is associated with disrupted immune functions such as immunosuppression and inflammatory suppression (Figure 3C-D).
Multiple Gene Co-Expression Modules Linked to Aberrant Immune Cell Activity in UC Patients
Additionally, differentially expressed genes (DEGs) in UC patients were identified based on expression data from the training dataset comprising 495 UC patients and 320 healthy controls. A total of 1084 DEGs (|log2FC|>0.585, P_value <0.05) were identified, among which 663 DEGs were differentially up-regulated and 421 DEGs were differentially down-regulated (Figure 4A). Subsequently, enrichment analysis exposed that the DEGs were enriched in the IL6 JAK STAT3 signaling pathway, neutrophil extracellular trap formation, KARS signaling, and other immune- and inflammation-related pathways (Figure 4B). Next, the correlations between DEGs and the abundance of immune cells were investigated. Six co-expression modules of DEGs were identified (MEgreen, MEblue, MEbrown, MEturquoise, MEyellow and MEgrey). Three of these modules were associated with aberrant immune cells in UC (|R| >0.4, P_value <0.05). Of note, the MEblue module was mainly positively associated with macrophages and neutrophils and negatively associated with CD4_T, CD4_naive, Tr1, iTreg, and Th2 cells. Meanwhile, MEturquoise was positively associated with CD4_naive, Tr1, iTreg and effector memory T cells, whereas MEgrey was positively correlated with macrophages and negatively correlated with Tr1 and iTreg cells (Figure 4C).
Construction of UC Diagnostic Model Based on Machine Learning
The Random Forest algorithm was applied to identify genes in the three immune cell-related modules. In the MEblue module, the Random Forest algorithm showed that the diagnostic error rate for UC was minimized with 47 model genes (Figure 5A). Afterward, genes in the MEblue module were ranked based on MeanDecreaseAccuracy, and the top 47 genes were selected for model construction (Figure 5B). Interestingly, the AUC of the 47-gene model for the diagnosis of UC was 0.862 in the training dataset (Figure 5C), 0.935 (Figure 5D) in the corresponding self-test dataset, and 0.732 (Figure 5E) in the independent test dataset, highlighting the diagnostic utility of the 47-gene model for UC. In the MEturquoise module, the machine learning algorithm was applied to develop diagnostic models, and optimal diagnostic efficacy was noted with 14 module genes (Figure 5F). Genes in the MEturquoise module were ranked based on MeanDecreaseAccuracy, and the top 14 genes were selected for model construction (Figure 5G). The results unveiled an AUC of 0.906 in the training dataset (Figure 5H), 0.900 in the self-test dataset, and 0.617 in the independent testing dataset (Figure 5I–J), highlighting the diagnostic efficacy of the top 14 genes in MEturquoise. Lastly, in the MEgrey module, the Random Forest algorithm identified the optimal model with 38 genes (Figure 5K), which were ranked based on MeanDecreaseAccuracy, following which the top 38 genes were selected for model construction (Figure 5L). The results uncovered an AUC of 0.938 in the training dataset (Figure 5M), 0.949 in the self-test dataset, and 0.662 in the independent testing dataset for the diagnosis of UC (Figure 5N–O), suggesting that the model possessed favorable diagnostic utility for UC.
Identification of the Optimal Diagnostic Biomarkers for UC
Based on the analysis of the random forest algorithm in the three gene modules, a total of 99 candidate genes with diagnostic efficacy in UC patients were identified. Subsequently, the Best Subset Selection regression model was applied to the candidate genes to screen the optimal diagnostic model for UC.
The results showed that the model had the largest R2 and adjusted R2 as well as the smallest Bayesian Information Criterion (BIC) and mallows_up values with 8 candidate genes, suggesting its optimal model efficacy (Figure 6A). The model consisted of 8 crucial genes (GATA2, IL8, LAT, NOLC1, SMARCA5, SMC3, STX10, and ZMIZ1) (Figure 6B). The AUC of this 8-gene model for the diagnosis of UC was 0.964 in the training dataset (Figure 6C), 0.926 in the self-test dataset (Figure 6D), and 0.884 in the independent testing dataset (Figure 6E). Indeed, the diagnostic efficacy of this 8-gene model significantly outperformed the above three diagnostic models.
Discussion
Ulcerative colitis (UC) and Crohn’s disease (CD), both classified as chronic inflammatory bowel diseases (IBD), are marked by alternating periods of exacerbation and remission.15 As is well documented, genetic, environmental, and immune factors significantly influence the pathogenesis of IBD.16 UC is characterized by continuous inflammation in the lamina propria of the colon, and its pathogenesis includes factors related to innate immunity, such as intestinal endothelial cell death, increased intestinal endothelial permeability,17 activation of proinflammatory M1-like macrophages, DC-induced proinflammatory immune responses,18 anti-inflammatory responses elicited by DC through induction of Tregs,19 increased proportions of NK cells,20 increased expression levels of defensins,21 and aberrant expression of PRRs such as TLR4.22 Noteworthily, its pathogenesis is also associated with adaptive immunity, such as the Th 1/Th 2 balance (in UC, the T-cell response to antigens is not Th 1-dominated but rather Th 2-dominated)23, an increase in the proportion of Th 17 cells24 and a decrease in the proportion of Tregs.25 Interestingly, the analysis of the datasets encompassing thousands of UC and control patients led to the identification of a distinct immune microenvironment associated with UC, characterized by the pivotal roles of immune cells such as Th2, Treg, and Th17. Additionally, machine learning algorithms assisted in the identification of eight immune microenvironment-associated biomarkers (GATA2, IL8, LAT, NOLC1, SMARCA5, SMC3, STX10, and ZMIZ1) with potential diagnostic efficacy for UC. Herein, GATA2 plays a decisive role in regulating the transcription of genes involved in the development and proliferation of hematopoietic and endocrine cell lines.26 It has been found to regulate dendritic cell differentiation,27 as well as being associated with macrophages.28 GSEA analysis of GATA2 in UC patients also demonstrated its association with Th17, Th1, and Th2 differentiation (Supplemental Figure 2). IL8, also referred to as CXCL8, plays a key role in mediating the inflammatory response and is secreted by various cell types, including monocyte macrophages, neutrophils, eosinophils, T lymphocytes, epithelial cells, and fibroblasts. It is worthwhile emphasizing that it is a chemokine that directs neutrophils to the site of infection and participates in pro-inflammatory signaling cascades with other cytokines.29 LAT activates the T cell antigen receptor (TCR) signaling pathway and is a key transport protein for activating essential amino acids required for immune responses in human T cells.30,31 NOLC1 enables molecular function inhibitor activity, is implicated in translational regulation, and has been identified as a tumor suppressor gene.32 SMARCA5 exhibits ATPase activity that regulates transcription by altering chromatin structure around genes.33 SMC3 regulates B cell transit through the germinal center.34 In the present study, we identified its association with the Spliceosome and chromatin remodeling processes in UC. (Supplemental Figure 2). STX10 encodes a protein involved in docking and fusion activities at the Golgi apparatus.35 Herein, GSEA revealed its role in the RIG-I-like receptor signaling pathway in UC (Supplemental Figure 2). ZMIZ1 regulates the activity of numerous transcription factors, including androgen receptor, Smad3/4, and p5336 and plays a critical role in T cell development.37 Although our study confirmed the upregulation of cytokines and provided transcriptomic evidence for the eight-gene diagnostic panel, protein-level validation of individual marker genes such as IL8 and GATA2 was not performed in this cohort due to tissue availability constraints. Future studies incorporating multiplex immunostaining or proteomic profiling will be essential to further verify their translational applicability in clinical settings.
Our diagnostic model integrates Random Forest (RF) feature importance with Best Subset Selection (BSS) to construct an interpretable eight-gene panel. RF allows for robust ranking of non-linearly associated features, while BSS ensures a parsimonious final model. The model performed well across internal and external datasets (AUC > 0.85), suggesting generalizability across populations. Unlike deep learning methods, our approach maintains clinical interpretability, making it more suitable for translational use. Compared to prior machine learning studies in UC that focused solely on differential expression, our method provides an immune-informed, pathway-grounded approach that may better reflect disease biology.
Moreover, the identified immune-related biomarkers may serve not only as diagnostic tools but also as potential targets for precision therapies. Advances in targeted delivery systems - such as antibody-drug conjugates, nanoparticles, or RNA-based platforms - offer the opportunity to deliver immunomodulatory agents directly to the inflamed intestinal tissue, potentially enhancing efficacy while minimizing systemic side effects. The integration of biomarker-based stratification with these delivery systems may represent a promising direction for future UC therapies and translational applications. In addition to direct immunological and transcriptomic markers, recent advances underscore the therapeutic relevance of microbiota-derived immunomodulators, such as short-chain fatty acids (SCFAs), indole derivatives, and bacterial extracellular vesicles. These metabolites influence key immune pathways including Treg/Th17 balance, inflammasome activation, and epithelial barrier integrity. Integrating host gene signatures with microbial metabolite profiles may enable co-targeting strategies that synergize immune diagnostics with microbial modulation.38 Future studies may investigate whether our identified biomarkers correlate with specific microbial signatures or functional metabolites, potentially guiding more personalized and mechanistically-informed treatment.
Translational Implications
The eight-gene diagnostic panel identified in this study not only facilitates molecular diagnosis of UC but may also provide a foundation for precision therapy. Several of the eight genes are involved in immune cell signaling and inflammation, making them promising targets or anchoring points for novel delivery strategies. Recent developments in reactive oxygen species (ROS) responsive nanocarriers and biomimetic cell membrane systems have enabled targeted modulation of inflammatory tissues with enhanced specificity and reduced systemic toxicity.14,39 In parallel, microbiota-derived immunomodulators such as short-chain fatty acids and tryptophan metabolites are being explored for their therapeutic synergy with immune-targeted approaches.38 Integrating transcriptomic biomarkers with these advanced platforms holds promise for achieving personalized, localized, and multimodal therapy in ulcerative colitis.
Limitations
This study has several limitations. First, detailed clinical information such as medication history and disease activity scores was incomplete in some public GEO datasets, which may introduce residual confounding despite our efforts to select well-annotated samples. In contrast, our local cohort included prospectively collected, treatment-naïve UC patients with complete clinical documentation, helping to mitigate this issue. Second, the IHC validation was performed on a relatively small cohort (24 UC and 21 controls) due to strict inclusion criteria. Although consistent with transcriptomic trends, larger cohorts and functional validation - including gene perturbation, in vitro models, and correlation with treatment outcomes - are needed to fully establish the clinical utility. Then, while the eight-gene diagnostic model showed strong performance in internal and external datasets, its AUC declined in the independent validation set. This may be due to overfitting or inter-dataset heterogeneity, including demographic differences, sampling protocols, and sequencing platforms. These findings highlight the need for multicenter validation and batch effect correction strategies. Immune cell estimation was based on bulk RNA-seq data, which averages signals across heterogeneous cell populations and may obscure subtle, spatially restricted immune dynamics. Future studies employing single-cell or spatial transcriptomics are warranted to refine immune landscape resolution. Lastly, immunomodulatory therapies (eg, corticosteroids, biologics, 5-ASA) can influence gene expression and immune profiles. While our local cohort was restricted to untreated patients, treatment status was incompletely annotated in public datasets. More comprehensive pharmacologic data will be crucial in future studies to isolate disease-intrinsic signatures.
Conclusion
In summary, this study highlights the significant role of immune microenvironment dysregulation in the pathogenesis of UC. Analyzing gene expression data and immune cell infiltration profiles from UC patients led to the identification of several aberrantly expressed immune cells, including B cells, macrophages, and various T cell subtypes, such as CD4+ naive T cells and Th2 cells. This immune imbalance is associated with the chronic inflammatory nature of UC. At the same time, gene co-expression analysis and machine learning models led to the identification of eight key biomarkers, namely GATA2, IL8, LAT, NOLC1, SMARCA5, SMC3, STX10, and ZMIZ1, which demonstrated strong diagnostic potential for UC. These findings provide novel insights into the immunopathogenesis of UC and offer promising targets for therapeutic intervention. Our results suggest that these biomarkers could facilitate early diagnosis and potentially guide the development of personalized treatments aimed at modulating the immune response in UC patients.
These findings provide novel insights into the immunopathogenesis of UC and offer promising targets for therapeutic intervention. Notably, several identified genes such as IL8, GATA2, and SMARCA5 are closely associated with inflammatory chemotaxis, immune activation, and epithelial barrier remodeling, positioning them as potential therapeutic targets. Beyond diagnostics, these genes may serve as molecular anchors for next-generation drug delivery systems. Recent advances in biomimetic nanotechnology, particularly cell membrane-coated nanoparticles and inflammation-targeting vesicles, have shown great potential for precision treatment of UC by improving drug localization and minimizing systemic side effects. For instance, cell membrane nanomaterials (CMNs) derived from immune or red blood cells exhibit immune evasion, tissue homing, and ROS-responsive release, making them ideal for integrating with biomarker-based targeting strategies.39 Future work may explore how our biomarkers can guide or synergize with these delivery systems to enable precise, personalized intervention in UC.
Abbreviations
IBD, Inflammatory bowel disease; UC, Ulcerative colitis; CD, Crohn’s disease; DCs, dendritic cells; Tregs, regulatory T cells; iTreg, induced regulatory T cell; MΦ, macrophages; Tr1, type 1 regulatory T cell; Tfh, T follicular helper cell; ECM, extracellular matrix; H&E, Hematoxylin-eosin; IHC, Immunohistochemistry; OD, optical density scoring; HPFs, high-power fields; DEGs, Differentially expressed genes; WGCNA, Weighted correlation network analysis; RF, Random forest; FDR, false discovery rate; BSS, Best Subset Selection; BIC, Bayesian Information Criterion; TCR, T cell antigen receptor.
Data Sharing Statement
The datasets used and analyzed during the current study are available in the GEO database under the accession IDs GSE177044 and GSE186507.
Ethics Approval
The current study met the requirements of the Declaration of Helsinki of the World Medical Association. Since this study did not require any intervention or experiment related to patients, no informed consent was required after review by the Ethics Committee of the First People’s Hospital of Yunnan Province. Ethical approval for FFPE samples in this study was obtained from the Ethics Committee of the First People’s Hospital of Yunnan Province (KHLL2024-KY199).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Open Foundation of Yunnan Provincial Laboratory of Clinical Virology (2023A4010403-02) and Open Foundation of Yunnan Digestive Endoscopy Clinical Medical Center (2022LCZXKF-XH18) and supported in part by the Health Commission Foundation of Yunnan Province (2023-KHRCBZ-B15), National Natural Science Foundation of China (No. 82404091), Yunnan Province Applied Basic Research Program Kunming Medical University Joint Project (202201AY070001-240), Basic Research Science and Technology Foundation of Yunnan Province (202201AS070009) and Xing Dian Foundation of Yunnan Province (XDYC-MY-2022-0029). Open Foundation of the First People’s Hospital of Yunnan Province (No. 2022LCZXKF-XB01 and No. 2023YJZX-HX01).
Disclosure
The authors declare that they have no competing interests.
References
1. Daniel K, Vitetta L, Fiatarone Singh MA. Effects of olives and their constituents on the expression of ulcerative colitis: a systematic review of randomised controlled trials. Br J Nutr. 2022;127(8):1153–1171. doi:10.1017/S0007114521001999
2. Xiong T, Zheng X, Zhang K, et al. Ganluyin ameliorates DSS-induced ulcerative colitis by inhibiting the enteric-origin LPS/TLR4/NF-kappaB pathway. J Ethnopharmacol. 2022;289:115001. doi:10.1016/j.jep.2022.115001
3. Choi D, Stewart AP, Bhat S. Ozanimod: a first-in-class Sphingosine 1-Phosphate receptor modulator for the treatment of ulcerative colitis. Ann Pharmacother. 2022;56(5):592–599. doi:10.1177/10600280211041907
4. Mansouri P, Mansouri P, Behmard E, Najafipour S, Kouhpayeh A, Farjadfar A. Novel targets for mucosal healing in inflammatory bowel disease therapy. Int Immunopharmacol. 2024;144:113544. doi:10.1016/j.intimp.2024.113544
5. Liao X, Liu J, Guo X, et al. Origin and function of monocytes in inflammatory bowel disease. J Inflamm Res. 2024;17:2897–2914. doi:10.2147/JIR.S450801
6. Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288(5474):2222–2226. doi:10.1126/science.288.5474.2222
7. Schurch CM, Bhate SS, Barlow GL, et al. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell. 2020;183(3):838. doi:10.1016/j.cell.2020.10.021
8. de Souza HS, Fiocchi C. Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol. 2016;13(1):13–27. doi:10.1038/nrgastro.2015.186
9. Eisenstein M. Gut reaction. Nature. 2018;563(7730):S34–S35. doi:10.1038/d41586-018-07277-1
10. Jones GR, Bain CC, Fenton TM, et al. Dynamics of colon monocyte and macrophage activation during colitis. Front Immunol. 2018;9:2764. doi:10.3389/fimmu.2018.02764
11. Chen S, Zeng J, Li R, et al. Traditional Chinese medicine in regulating macrophage polarization in immune response of inflammatory diseases. J Ethnopharmacol. 2024;325:117838. doi:10.1016/j.jep.2024.117838
12. Yang FC, Chiu PY, Chen Y, Mak TW, Chen NJ. TREM-1-dependent M1 macrophage polarization restores intestinal epithelium damaged by DSS-induced colitis by activating IL-22-producing innate lymphoid cells. J Biomed Sci. 2019;26(1):46. doi:10.1186/s12929-019-0539-4
13. Ungaro R, Mehandru S, Allen PB, Peyrin-Biroulet L, Colombel JF. Ulcerative colitis. Lancet. 2017;389(10080):1756–1770. doi:10.1016/S0140-6736(16)32126-2
14. Wan X, Zhang C, Lei P, et al. Precision therapeutics for inflammatory bowel disease: advancing ROS-responsive nanoparticles for targeted and multifunctional drug delivery. J Mater Chem B. 2025;13(10):3245–3269. doi:10.1039/d4tb02868f
15. Sosna B, Aebisher D, Mysliwiec A, et al. Selected cytokines and metalloproteinases in inflammatory bowel disease. Int J Mol Sci. 2023;25(1). doi:10.3390/ijms25010202
16. Tsuda S, Carreras J, Kikuti YY, et al. Prediction of steroid demand in the treatment of patients with ulcerative colitis by immunohistochemical analysis of the mucosal microenvironment and immune checkpoint: role of macrophages and regulatory markers in disease severity. Pathol Int. 2019;69(5):260–271. doi:10.1111/pin.12794
17. Gassler N, Rohr C, Schneider A, et al. Inflammatory bowel disease is associated with changes of enterocytic junctions. Am J Physiol Gastrointest Liver Physiol. 2001;281(1):G216–28. doi:10.1152/ajpgi.2001.281.1.G216
18. Drakes ML, Blanchard TG, Czinn SJ. Colon lamina propria dendritic cells induce a proinflammatory cytokine response in lamina propria T cells in the SCID mouse model of colitis. J Leukoc Biol. 2005;78(6):1291–1300. doi:10.1189/jlb.0605342
19. Darrasse-Jeze G, Deroubaix S, Mouquet H, et al. Feedback control of regulatory T cell homeostasis by dendritic cells in vivo. J Exp Med. 2009;206(9):1853–1862. doi:10.1084/jem.20090746
20. Poggi A, Benelli R, Vene R, et al. Human gut-associated natural killer cells in health and disease. Front Immunol. 2019;10:961. doi:10.3389/fimmu.2019.00961
21. Rahman A, Fahlgren A, Sitohy B, et al. Beta-defensin production by human colonic plasma cells: a new look at plasma cells in ulcerative colitis. Inflamm Bowel Dis. 2007;13(7):847–855. doi:10.1002/ibd.20141
22. Franchimont D, Vermeire S, El Housni H, et al. Deficient host-bacteria interactions in inflammatory bowel disease? The toll-like receptor (TLR)-4 Asp299gly polymorphism is associated with Crohn’s disease and ulcerative colitis. Gut. 2004;53(7):987–992. doi:10.1136/gut.2003.030205
23. Tatiya-Aphiradee N, Chatuphonprasert W, Jarukamjorn K. Immune response and inflammatory pathway of ulcerative colitis. J Basic Clin Physiol Pharmacol. 2018;30(1):1–10. doi:10.1515/jbcpp-2018-0036
24. Jiang P, Zheng C, Xiang Y, et al. The involvement of TH17 cells in the pathogenesis of IBD. Cytokine Growth Factor Rev. 2023;69:28–42. doi:10.1016/j.cytogfr.2022.07.005
25. Mohammadnia-Afrouzi M, Zavaran Hosseini A, Khalili A, Abediankenari S, Hosseini V, Maleki I. Decrease of CD4(+) CD25(+) CD127(low) FoxP3(+) regulatory T cells with impaired suppressive function in untreated ulcerative colitis patients. Autoimmunity. 2015;48(8):556–561. doi:10.3109/08916934.2015.1070835
26. Bresnick EH, Jung MM, Katsumura KR. Human GATA2 mutations and hematologic disease: how many paths to pathogenesis? Blood Adv. 2020;4(18):4584–4592. doi:10.1182/bloodadvances.2020002953
27. Onodera K, Fujiwara T, Onishi Y, et al. GATA2 regulates dendritic cell differentiation. Blood. 2016;128(4):508–518. doi:10.1182/blood-2016-02-698118
28. Luo X, Meng C, Zhang Y, et al. MicroRNA-21a-5p-modified macrophage exosomes as natural nanocarriers promote bone regeneration by targeting GATA2. Regen Biomater. 2023:
29. Matsushima K, Yang D, Oppenheim JJ. Interleukin-8: an evolving chemokine. Cytokine. 2022;153:155828. doi:10.1016/j.cyto.2022.155828
30. Carpier JM, Zucchetti AE, Bataille L, et al. Rab6-dependent retrograde traffic of LAT controls immune synapse formation and T cell activation. J Exp Med. 2018;215(4):1245–1265. doi:10.1084/jem.20162042
31. Hayashi K, Jutabha P, Endou H, Sagara H, Anzai N. LAT1 is a critical transporter of essential amino acids for immune reactions in activated human T cells. J Immunol. 2013;191(8):4080–4085. doi:10.4049/jimmunol.1300923
32. Zhai F, Li Y, Luo X, Jin X, Ye M. NOLC1 was identified as a tumor suppressor gene in thyroid cancer and correlated with prognosis by bioinformatics. Am J Cancer Res. 2024;14(5):2055–2071. doi:10.62347/IYVV7581
33. Jevtic Z, Matafora V, Casagrande F, et al. SMARCA5 interacts with NUP98-NSD1 oncofusion protein and sustains hematopoietic cells transformation. J Exp Clin Cancer Res. 2022;41(1):34. doi:10.1186/s13046-022-02248-x
34. Rivas MA, Meydan C, Chin CR, et al. Smc3 dosage regulates B cell transit through germinal centers and restricts their malignant transformation. Nat Immunol. 2021;22(2):240–253. doi:10.1038/s41590-020-00827-8
35. Ganley IG, Espinosa E, Pfeffer SR. A syntaxin 10-SNARE complex distinguishes two distinct transport routes from endosomes to the trans-Golgi in human cells. J Cell Biol. 2008;180(1):159–172. doi:10.1083/jcb.200707136
36. Lomeli H. ZMIZ proteins: partners in transcriptional regulation and risk factors for human disease. J Mol Med. 2022;100(7):973–983. doi:10.1007/s00109-022-02216-0
37. Wang Q, Yan R, Pinnell N, et al. Stage-specific roles for Zmiz1 in Notch-dependent steps of early T-cell development. Blood. 2018;132(12):1279–1292. doi:10.1182/blood-2018-02-835850
38. Chen H, Lei P, Ji H, et al. Escherichia coli Nissle 1917 ghosts alleviate inflammatory bowel disease in zebrafish. Life Sci. 2023;329:121956. doi:10.1016/j.lfs.2023.121956
39. Lei P, Yu H, Ma J, et al. Cell membrane nanomaterials composed of phospholipids and glycoproteins for drug delivery in inflammatory bowel disease: a review. Int J Biol Macromol. 2023;249:126000. doi:10.1016/j.ijbiomac.2023.126000
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
Recommended articles
Serological Biomarker-Based Machine Learning Models for Predicting the Relapse of Ulcerative Colitis

Pang W, Zhang B, Jin L, Yao Y, Han Q, Zheng X
Journal of Inflammation Research 2023, 16:3531-3545
Published Date: 21 August 2023
Role of Aging in Ulcerative Colitis Pathogenesis: A Focus on ETS1 as a Promising Biomarker

Ni M, Peng W, Wang X, Li J
Journal of Inflammation Research 2025, 18:1839-1853
Published Date: 6 February 2025
Identification of Senescence-Related Genes for the Prediction of Ulcerative Colitis Based on Interpretable Machine Learning Models
Ma J, Chen C, Wang N, Fang T, Liu Y, He P, Dong W
Journal of Inflammation Research 2025, 18:3431-3447
Published Date: 10 March 2025
Identification and Validation of Feature Genes Related to Mitochondrial Dysfunction and Oxidative Stress in Ulcerative Colitis
Wu Y, Tan Z, Lan Q, Liu Y, Liu C, He H, Zhang J, Dong W
Journal of Inflammation Research 2025, 18:5835-5850
Published Date: 30 April 2025






