Back to Journals » Journal of Inflammation Research » Volume 17
MiRNAs and Neutrophil-Related Membrane Proteins from Plasma-Derived Extracellular Vesicles for Early Prediction of Organ Dysfunction and Prognosis in Septic Patients
Authors Ye R, Wei Y, Li J, Xu M, Xie H, Huang J, Deng L , Li C
Received 24 August 2024
Accepted for publication 28 November 2024
Published 4 December 2024 Volume 2024:17 Pages 10347—10369
DOI https://doi.org/10.2147/JIR.S492902
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Tara Strutt
Rongzong Ye,1,* Yating Wei,1,* Jingwen Li,2,* Meili Xu,1 Haiyang Xie,2 Jiahao Huang,3 Liehua Deng,3 Chaoqian Li1
1Department of Emergency Medicine, The First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi, 530021, People’s Republic of China; 2Guangxi Medical University, Nanning, Guangxi, 530021, People’s Republic of China; 3Department of Critical Care Medicine, Affiliated Hospital of Guangdong Medical University, Zhanjiang, Guangdong, 524000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Liehua Deng, Department of Critical Care Medicine, Affiliated Hospital of Guangdong Medical University, 57 People’s Ave South, Zhanjiang, Guangdong, 524000, People’s Republic of China, Email [email protected] Chaoqian Li, Department of Emergency Medicine, The First Affiliated Hospital of Guangxi Medical University, 6 Shuangyong Road, Nanning, Guangxi, 530021, People’s Republic of China, Email [email protected]
Purpose: The pathogenesis of sepsis-induced organ dysfunction remains elusive, and the mortality remains alarmingly high. We sought to investigate the profile of extracellular vesicles (EVs)-mediated communication between plasma and polymorphonuclear neutrophils (PMNs) in sepsis, and to elucidate whether miRNAs and PMN-related membrane proteins from plasma-derived EVs (plasma-EVs) are associated with sepsis-induced organ dysfunction and prognosis.
Methods: PMN-derived EVs (PMN-EVs) were isolated from the blood samples of healthy controls (N=3) and patients with septic shock (N=3) after ICU admission. We performed miRNA sequencing of the isolated EVs, followed by bioinformatic analysis. A miRNA model for comparing PMN-EVs and plasma-EVs was successfully established in the training cohort. Furthermore, miRNAs and PMN-related membrane proteins from the plasma-EV model were confirmed in the validation cohort. A logistic regression model, receiver operating characteristic (ROC) curves, and Kaplan-Meier analyses were performed to evaluate the efficiency of diagnostic and/or prognostic performance. Further, in vivo and in vitro experiments were conducted to explore the involvement of plasma-EVs in PMNs autophagy.
Results: Fifty-five miRNAs from PMN-EVs differed significantly between the healthy controls and patients with septic shock. Furthermore, the plasma-EV model (six miRNAs and eight PMN-related membrane proteins) was confirmed in the validation cohort, demonstrating that miR-34a-5p, miR-503-5p, miR-4772-3p, ITGAM, MPO, and MMP9 serve as sepsis biomarkers for distinguishing lung, liver, and kidney dysfunction. Kaplan–Meier survival analysis showed that miR-34a-5p, miR-4772-3p, ITGAM, and MMP9 were potential prognostic predictors. Finally, we found that plasma-EVs from sepsis patients exert an inhibitory effect on PMNs autophagy, which can be reversed by EV inhibitors such as GW4869 and enoxaparin.
Conclusion: These findings suggest that miRNAs and PMN-related membrane proteins from plasma-EVs could be valuable diagnostic tools for identifying sepsis-induced organ dysfunction and predicting prognosis, enabling proactive management of sepsis by physicians and improving the prognosis of sepsis patients.
Keywords: extracellular vesicles, MicroRNAs, membrane proteins, sepsis-induced organ dysfunction, neutrophils, autophagy
Introduction
Sepsis, a life-threatening organ dysfunction triggered by severe infection, remains a prominent cause of morbidity and mortality worldwide.1 Regrettably, many patients with sepsis-induced multiple organ dysfunctions experience higher mortality rates primarily due to the fact that the pathophysiology, effective classification diagnostic tools, and novel therapeutic strategies in sepsis remain not fully elucidated.2–6
Extracellular vesicles (EVs) exhibit high specificity as they contain a diverse array of proteins, RNA, and other molecules that directly originates from their parent cells and facilitate intercellular communications.7,8 This nudges EVs to accurately reflect the physiological and pathological states of these cells, making them not only potential biomarkers for the early diagnosis and prognosis of sepsis but also opening avenues for novel therapeutic strategies. The serum exosomal miRNAs demonstrated superior performance compared to routine parameters in accurately predicting persistent organ failure, and could forecast the onset of persistent organ failure within the “golden hours” following acute pancreatitis.9 A study on the plasma exosomal proteomics analysis of individuals with sepsis-induced acute lung injury (ALI) and healthy controls revealed differential expression of 39 proteins. Additionally, elevated levels of plasma exosomal APN/CD13 were correlated with the severity of ALI and mortality in sepsis patients.10 Moreover, plasma-derived EVs are able to cross the blood-brain barrier in normal mice, and in sepsis they can induce neuronal inflammation in the brain through miRNAs and innate immune signaling.11 Therefore, circulating EVs may play a crucial role in modulating intercellular communication between immunological and structural cells, as well as contributing to sepsis-induced organ dysfunction.
Recent studies have demonstrated a clear link between the potent antibacterial components of polymorphonuclear neutrophils (PMNs) and the emergence of organ dysfunction in sepsis.12 In previous studies, EVs generated by the donor cells were stimulated in vitro by LPS, which may not fully reflect the intricate in vivo scenario during sepsis; however, there are few reports on the characteristic profile of EVs in PMNs from early septic patients, as well as EV-mediated signal transmission between plasma and PMNs. In light of these considerations, we aimed to elucidate whether specific miRNAs and membrane proteins from plasma-EVs are associated with particular sepsis-induced organs dysfunction and prognosis. Furthermore, we sought to unravel new hints for researches on EVs in PMNs and plasma in sepsis, as well as provide insights for clinical practitioners.
Materials and Methods
Patients and Control Subjects
Patients with sepsis admitted to the Emergency Medicine of the First Affiliated Hospital of Guangxi Medical University from January to December 2023 were included as the study subjects, and healthy volunteers were recruited as the control group. Sepsis was diagnosed based on the guidelines of the Third International Consensus Deficiency for Sepsis and Septic Shock (Sepsis-3). Sepsis-induced organ dysfunction was diagnosed retrospectively according to the Criteria to Screen for Sepsis or Assess Organ Dysfunction.13 The discovery cohort consisted of 3 patients with septic shock at admission (day 1 and day 3) and 3 healthy controls. The study enrolled a total of 96 patients diagnosed with sepsis, among whom 7 were excluded due to insufficient samples for ELISA analysis, and 5 were lost to follow-up, resulting in a final enrollment of 84 patients in the validation cohort. Additionally, 18 healthy donors were enrolled as controls, with an average age of 35±11, including 10 males (55.6%) and 8 females (44.4%). Baseline demographics and clinical details of the subjects were retrospectively collected. We excluded patients who were pregnant, had severe anemia or active bleeding, neutropenia, a history of congenital heart disease, coronary heart disease, myocardial infarction, hypertensive heart disease, pulmonary hypertension, chronic heart dysfunction, tumors or organ transplants, an immunosuppressive or immune-deficient state, and those who were not reachable during follow-up. Peripheral blood samples of 20 mL were collected from patients in EDTA tubes following a standard venipuncture procedure. After centrifugation at 3000 ×g for 15 min at 4°C, the plasma was aspirated and stored at −80°C. All protocols, procedures, and subject/patient recruitment procedures described in this study were approved by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University, China (Approval No.2023-K128-01). This study was conducted in accordance with the principles of the Declaration of Helsinki. Written informed consent was obtained from all the subjects and patients.
Animals
Adult male Sprague-Dawley rats (6–8 weeks old) were purchased from the Animal Center of Guangxi Medical University and were maintained under specific pathogen-free conditions. All animal experiments were conducted in accordance with the National Institutes of Health “Guidelines for Care and Use of the Laboratory Animals” and approved by the Animal Ethics Committee of Guangxi Medical University (Approval No. 202305565). Every effort was made to minimize the number of rats used and their suffering.
PMNs Isolation
Peripheral blood samples were collected from humans and rats. PMNs were isolated using a Peripheral Blood Neutrophil Isolation Reagent Kit (Solarbio), according to the manufacturer’s instructions. Briefly, 40 mL peripheral blood was collected in a tube containing an anticoagulant and mixed slowly with reagents A and C, followed by centrifugation at 1000 ×g for 20 min. Following centrifugation, the neutrophil-containing layer between reagents A and C was gently removed and lysed with Red Blood Cell Lysis Buffer to remove the erythrocytes. Healthy donors or septic patients PMNs were cultured in 1640 medium containing 10% EVs-free FBS at 37 °C and 5% CO2 for 24 hours, and then the culture supernatant was collected to isolate EVs.
EVs Purification and Characterization from Plasma and PMNs
EVs were isolated by ultracentrifugation (UC) and molecular size exclusion (SEC). For the plasma samples, 20 mL peripheral blood samples from patients were collected in EDTA tubes following a regular venipuncture procedure. The collected blood samples should be promptly centrifuged within 2 hours to remove cellular debris and stored at −80°C for subsequent extraction of EVs. In short, the plasma should be thawed at 37°C and then centrifuged in 3-steps: 1500 ×g for 20 min at 4°C; 3000 ×g for 15 min at 4°C; 13,000×g for 30 min at 4°C, and processed through a 0.22 μm filter to remove large particles. For the PMNs medium, PMNs were cultured in 10% EVs-free FBS medium for 24 hours. In short, 3-step centrifugation at 300 ×g for 10 min; 3000 ×g for 10 min at 4°C and processed through a 0.22 μm filter to remove large particles. The supernatant of plasma or PMNs was then ultracentrifuged using a Type70Ti (Beckman, USA) at 100,000 ×g for 2 hours at 4°C to pellet the EVs. Then, the pellet was re-suspended in PBS and centrifuged at 100,000 ×g for 2 hours at 4°C. After washing with PBS, the EVs pellet was re-suspended in 1mL PBS. SEC was used to separate EVs using an Exosupur exclusion column (Echobiotech, China), following the manufacturer’s instructions. The sample was then eluted with PBS and 2 mL of the target fraction was collected. Finally, the EVs solution was concentrated using an Amicon® ultrafiltration tube with a molecular weight cutoff of 100kDa (Merck, Germany). It was divided into two portions, one used for ELISA detection and the other for RNA extraction.
EV morphology was examined under transmission electron microscope (HT-7700, Hitachi Ltd, Japan). EVs surface marker proteins were resolved by SDS-polyacrylamide gel electrophoresis and immunoblotted using antibodies against the following proteins: ALIX (1:1000, R23425, Zen-bio), CD9 (1:1000, 380441, Zen-bio), TSG101 (1:1000, 381538, Zen-bio), Calnexin (1:5000, 10,427-2-AP, Proteintech), VDAC1 (1:1000, 55,259-1-AP, Proteintech), GM130 (1:5000,11,308-1-AP, Proteintech), and TIM23 (1:1000, 11,123-1-AP, Proteintech). The size distribution and particle concentration of EVs were characterized by Nano flow cytometry (NanoFCM).
RNA Sequencing and miRNA Analysis
RNA sequencing and differential expression analyses were performed as previously described.14 miRDB and miRWalk were used to predict the biological targets of differentially expressed miRNAs (DE-miRNAs), and those that overlapped with the targets of differentially expressed genes were identified. Gene functions and signaling pathways were annotated using the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases. The UniProt database indicates the presence of 24 exosome membrane proteins associated with neutrophils. The TRRUST database was used to predict the TF-mRNA relationships. The TF-miRNA-mRNA network was constructed and visualized using Cytoscape (version 3.10), based on the regulatory relationship between TFs, miRNAs, and mRNAs.
RNA Extraction and Quantitative Real-Time PCR
Total RNA was extracted from plasma or PMNs derived EVs using an Exosome RNA Purification Kit (5202050, Simgen). RNA was converted into complementary DNA (cDNA) using the Mir-XTM miRNA First-Strand Synthesis Kit (638315, TaKaRa). qPCR analysis was performed with TB Green Advantage qPCR Premix (RR820A, TaKaRa). RT-qPCR analysis was performed using the Applied Biosystems system. U6 served as an internal control to normalize the qPCR data. The experiments were conducted in triplicate, and the relative fold changes in miRNA expression levels were calculated using the following equation: relative quantification = 2–ΔΔCT. Primers used are listed in Supplementary Table 1.
Enzyme-Linked Immunosorbent Assay
PMN-related membrane proteins in plasma-EVs were quantified using MPO (SEA601Hu, Cloud-Clone Corp)., MMP9 (SEA553Hu, Cloud-Clone Corp)., ITGAM (SEB685Hu, Cloud-Clone Corp)., CXCR4 (SEA940Hu, Cloud-Clone Corp)., CXCR2 (SEC006Hu, Cloud-Clone Corp)., CSF3R(SEB476Hu, Cloud-Clone Corp)., CD33 (ELK9595, ELK Biotechnology), and SELL (ELK5204, ELK Biotechnology). The levels of tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β), and interleukin-6 (IL-6) in the serum samples were quantified using ELISA kits from Cloud-Clone Corp., China, according to the provided protocols. A microplate reader was used to measure the OD density at 450 nm. The concentrations of proteins in the EVs or serum were determined by comparing the OD of the samples to the standard curve.
Immunoblotting Analysis
Total protein was extracted using RIPA lysis buffer containing protease inhibitors. The protein concentrations in the supernatants were measured using a BCA assay kit (P0010, Beyotime). The proteins were separated by SDS-PAGE and transferred to PVDF membrane (Millipore). Membranes were blocked in TBST containing 5% bovine serum albumin at room temperature for 1 hours, and incubated with primary antibodies overnight at 4°C. β-actin was used as an internal control. The following antibodies were used: ATG5 (1:1000, R23497, Zen-bio), Beclin1 (1:1000, R22856, Zen-bio), P62 (1:1000, 380612, Zen-bio), CXCR4 (1:1000, 380981, Zen-bio), MMP9 (1:1000, 380831, Zen-bio), LC3B (1:1000, T55992S, Abmart), CXCR2 (1:1000, ab65968, Abcam), ITGAM (1:1000, ab133357, Abcam), ATG7 (1:2000, CY7051, Abways), and β-actin (1:10,000, 380624, Zen-bio). After washing with TBST, the membrane was incubated with the Anti-rabbit IgG (H+L) (DyLight™ 800 4X PEG Conjugate) (1:20,000, 5151, CST) for 1 hours at room temperature. After washing with TBST, images were captured by Odyssey software.
EV Fluorescence Labelling
To investigate whether plasma-EVs were taken up by PMNs, they were labeled with the fluorescent lipophilic tracer DiR before co-culture. Labeling was performed using a DiR-label Kit (D-9111, Bioss), following the manufacturer’s instructions. Briefly, the EVs solution was mixed with the staining solution (2 µM), vigorously shaken for 1 min, and incubated for 20 min. Finally, all the unbound dye molecules were removed by UC at 100,000 × g for 90 min at 4°C and the pellets were resuspended in PBS. After co-culture for 6 hours, PMNs were fixed, stained with DAPI (C02-04002, Bioss), and observed under an Olympus fluorescence microscope (Olympus, Japan).
In vitro Co-Culture of EVs Experiments
Isolated EVs were quantified using the BCA assay. In vitro, human PMNs were co-cultured with 100 μg/mL plasma-EVs for 24 hours from healthy donors (Control-EVs) or septic patients (Sepsis-EVs).
LPS-Induced Sepsis Model
To induce experimental sepsis, rats were treated with either an intraperitoneal injection of endotoxin lipopolysaccharides (Escherichia coli O111:B4, Sigma Aldrich) 10 mg/kg body weight or saline and sacrificed after 24 hours. Rat blood was collected in 5mL EDTA tubes and EVs were isolated according to a previously described protocol.
Treatment of EVs Inhibitors in Septic Rat
The EV-release inhibitor GW4869 and EV-uptake inhibitor enoxaparin were used to evaluate the role of plasma-EVs in sepsis development. Briefly, rats were intraperitoneally administered GW4869 (2.5 mg/kg) 1 hours before LPS induction. In another experiment, enoxaparin (3 mg/kg) was injected into healthy rats through the tail vein, followed by the injection of LPS-EVs or Saline-EVs (600 μg in 100 μL PBS per rat). Rats injected with the same volume of saline were used as controls. The rats were euthanized 24 hours after injection, and peripheral blood was collected in EDTA tubes to isolate PMNs for further analysis.
Statistical Analysis
The Shapiro–Wilk test was performed to test the normality of the data. The baseline characteristics of all patients conforming to a normal distribution are expressed as mean±standard deviation (SD). The results for miRNAs and membrane proteins are expressed as the mean ± standard error of the mean (SEM). For non-normally distributed data, continuous variables were reported as medians and interquartile ranges, and categorical variables were reported as numbers and percentages. Comparisons of continuous and categorical variables between the groups were performed using the Mann–Whitney U-test, Fisher’s exact test, or chi-square test, respectively. Unpaired Student’s t-test was used to analyze the differences between two groups, and differences among three or more groups were analyzed using one-way analysis of variance (ANOVA) or two-way ANOVA, followed by a post hoc Tukey’s test. Correlation analyses were performed using the Spearman correlation coefficient. A logistic regression model was employed to compute hazard ratios and accompanying 95% confidence intervals. Receiver operating characteristic (ROC) curve analysis was used to evaluate the efficiency of the diagnostic and/or prognostic performance. To graphically illustrate the impact of miRNAs or membrane proteins, the data were dichotomized based on low versus high expression at the median. Survival rates between groups were compared using the Kaplan-Meier method and the Log rank test. All statistical analyses were performed using SPSS software (version 26.0) and GraphPad Prism version 9.0. Statistical significance was set at P < 0.05.
Results
miRNA Profiles from PMN-EVs in Septic Shock Patients
To clarify whether miRNAs from PMN-EVs are key mediators of organ dysfunction in septic shock, sequencing of PMN-EVs was performed to identify potentially efficient miRNAs. We then screened 55 differentially expressed miRNAs (DE-miRNAs), of which 9 were downregulated and 46 were upregulated. The clustering heatmap visually displayed these DE-miRNAs (Supplementary Table 2) (Figure 1A). We then utilized the miRWalk and miRDB databases to predict the target genes for the 55 DE-miRNAs, which collectively had 3863 target genes. GO analysis revealed that 1978 significantly enriched GO terms encompassing 1544 biological processes, 215 cellular components, and 219 molecular functions were involved in the regulation of cell morphogenesis, cell–cell junction, and transcriptional coregulator activity, etc (Figure 1B). Meanwhile, 141 significantly enriched pathways were obtained by KEGG analysis, mainly involving the MAPK signaling pathway, Ras signaling pathway, and autophagy, etc (Figure 1C). Heatmap display showing 11 differentially expressed miRNAs in patients with septic shock at day 1 and day 3 (Figure S1A) (Supplementary Table 3). Then, we utilised miRWalk and miRDB databases to predict the target genes for DE-miRNAs, which collectively had 1334 target genes in both databases. KEGG and GO analyses were used to assess the potential functions of these DE-miRNAs based on their target genes (Figure S1B and C).
To explore miRNAs and PMN-related surface proteins that target the interaction regulatory network, we used the UniProt database to identify 24 PMN-related membrane proteins. Twenty-four PMN-related target genes of the identified miRNA signatures were obtained from the miRWalk database. A regulatory network of TF-miRNA-mRNA was established, involving 16 transcription factors, 27 upregulated miRNAs (19 miRNAs were deleted because they did not participate in the network), and 17 PMN-related target genes (Figure 1D). Another TF-miRNA-mRNA network consisted of 3 transcription factors, 8 downregulated miRNAs (hsa-miR-10b-5p was deleted because they did not participate in the network), and 14 PMN-related target genes (Figure 1E). These results suggest that miRNAs derived from PMN-EVs may reflect and/or regulate sepsis progression by influencing multiple signalling pathways.
Screening DE-miRNAs from PMN-EVs in Plasma-EVs
EVs were isolated from the plasma or PMNs medium of patients with septic shock by ultracentrifugation and molecular size exclusion. Transmission electron microscopy (TEM) images showed that the morphology of the PMN-derived or plasma-derived EVs was typical of Exos, as round bulging structures were observed (Figure 2A). All isolated EVs showed a similar size distribution profile with isolated particles within the 30–150 nm range by NanoFCM analysis. The concentration of isolated PMN-derived or plasma-derived EVs was approximately 1.51E+8 particles/mL, and 1.79E+8 particles/mL respectively (Figure 2B). Immunoblotting analysis showed that the isolated EVs expressed positive EVs biomarkers (Alix, Tsg101, and CD9), whereas Golgi (GM130), endoplasmic reticulum (Calnexin), and mitochondrial proteins (VDAC1 and TIM23) were exclusively detected in cell lysates, serving as negative markers for EVs (Figure 2C). These results demonstrate that PMNs and plasma secreted EVs with characteristics consistent with those of Exos.
To verify the differential expression of candidate miRNAs derived from PMN-EVs in the discovery phase, the expression of 13 miRNAs was detected using RT-qPCR in an independent training set consisting of 12 patients with septic shock and 12 healthy controls. The results revealed a significant difference and consistent expression trend between patients with septic shock and healthy controls in the expression of 9 candidate miRNAs (miR-34a-5p, miR-96-5p, miR-182-5p, miR-199b-5p, miR-210-3p, miR-301a-3p, miR-503-5p, miR-1262, and miR-4772-3p), as observed in the training set (Figure 3A). We hypothesized that PMN-EVs might enter the bloodstream during sepsis. Therefore, we conducted a comparative analysis of miRNA expression in PMN-EVs and plasma-EVs obtained from the same patient with sepsis. The results of the differential expression analysis of 9 miRNAs in PMN-EVs were used as a basis for further verification to confirm their differential expression in plasma-EVs. The results showed that the expression of miR-301a-3p in the plasma-EVs of septic shock patients was significantly lower than that in the control group, whereas the expression of miR-34a-5p, miR-199b-5p, miR-210-3p, miR-503-5p, miR-1262, and miR-4772-3p was significantly higher (Figure 3B). Paired analysis of miRNA expression also indicated that miR-34a-5p, miR-199b-5p, miR-210-3p, miR-503-5p, and miR-4772-3p are higher in plasma-EVs. Compared to PMN-EVs, the level of miR-301a-3p was significantly lower in plasma-EVs, whereas the difference in the level of miR1262 did not reach statistical significance (Figure 3C).
Validation of DE-miRNAs and PMN-Related Membrane Proteins from Plasma-EVs
The baseline characteristics of study participants at admission are shown in Table 1. The median age was 58 (46–64.5) and 75% of the patients were male. There were no significant differences in age, sex, or comorbidities between the sepsis and septic shock groups, whereas the clinical characteristics, including pulmonary infection, PaO2/FiO2, MAP, lactate, CRP, albumin, serum creatinine, SOFA score, APACHE II score, and 28-day mortality, were significantly different (P<0.05).
 |
Table 1 The Baseline Characteristics of All the Patients |
We further validated the upregulation of 6 miRNAs (miR-34a-5p, miR-199b-5p, miR-210-3p, miR-503-5p, miR-1262, and miR-4772-3p) in plasma-EVs in an independent validation study consisting of 36 sepsis patients, 48 septic shock patients and 18 healthy controls. The levels of miR-34a-5p, miR-199b-5p, and miR-503-5p were significantly increased in sepsis patients compared with the healthy controls (P<0.05). In addition, the levels of miR-34a-5p, miR-199b-5p, miR-503-5p, and miR-4772-3p were significantly increased in septic shock compared to healthy controls and sepsis (Figure 4A).
We next validated 8 PMN-related membrane proteins from plasma-EVs, using a commercially available ELISA. The proteins CD33, CSF3R, CXCR2, CXCR4, ITGAM, MPO, MMP9, and SELL were selected for comparison. Compared to the healthy controls group, levels of ITGAM and MPO were found to be elevated in plasma-EVs in patients with sepsis; and the expression of CXCR2, ITGAM, MPO, and MMP9 was significantly higher in patients with septic shock. In addition, the levels of ITGAM, MPO, and MMP9 were higher in patients with septic shock than those with sepsis (Figure 4B).
To further evaluate potential biomarkers for predicting septic organ dysfunction, we also compared the differential expression of miRNAs and PMN-related membrane proteins in plasma-EVs. The demographic and clinical characteristics of the patients analyzed according to subgroup are provided in Supplementary Tables 4-7. The levels of miR-34a-5p, miR-503-5p, and MMP9 were significantly upregulated in the ARDS group, compared with Non-ARDS. The level of MPO was significantly elevated in the ALF group, as compared to Non-ALF. We found that miR-199b-5p, miR-4772-3p, MPO, and ITGAM levels tended to be higher in AKI group compared to Non-AKI (Figure 5A). We further validated these findings in prognosis, and the results showed that levels of miR-34a-5p, miR-4772-3p, ITGAM, and MMP9 in plasma-EVs of Non-survivors were significantly higher than those in the survivors group (Figure 5B). Taken together, our data suggest that the plasma-EVs panel (miR-34a-5p, miR-503-5p, miR-4772-3p, ITGAM, MPO, and MMP9) may be a potential noninvasive biomarker for septic organ dysfunction.
Association of miRNAs and PMN-Related Membrane Proteins to Organ Dysfunction and Prognosis in Septic Patients
We performed Spearman correlation analysis in the validation sets to assess whether the expression levels of the plasma-EV panel (miR-34a-5p, miR-503-5p, miR-4772-3p, ITGAM, MPO, and MMP9) correlated with severity. Notably, the expression levels of miR-34a-5p, miR-199b-5p, miR-4772-3p, ITGAM, MPO, and MMP9 were positively correlated with SOFA scores, whereas miR-503-5p was not correlate with SOFA scores (Figure S2A). In addition, Spearman correlation analysis showed that the expression levels of miR-34a-5p, miR-199b-5p, miR-4772-3p, ITGAM, MPO, and MMP9 were positively correlated with the APACH II score in patients with sepsis, but there was no such correlation for miR-503-5p (Figure S2B).
Next, we used multivariate logistic regression analysis to determine the best combination of miRNAs and PMN-related membrane proteins for predicting septic organ dysfunction and prognosis. The results show that independent predictors of sepsis-induced acute respiratory distress syndrome (S-ARDS) included miR-34a-5p (OR=1.931, 95% CI 1.104–3.376, P=0.021), miR-503-5p (OR=1.582, 95% CI 1.091–2.293, P=0.016), and MMP9 (OR=1.856, 95% CI 1.277–2.696, P=0.001). MPO (OR=1.018, 95% CI 1.010–1.026, P<0.001) and ITGAM (OR=1.006, 95% CI 1.002–1.010, P=0.002) emerged as independent predictors of sepsis-induced acute liver failure (S-ALF). Furthermore, high miR-4772-3p expression was an independent predictor of sepsis-induced acute kidney injury (S-AKI) (OR=2.639, 95% CI 1.503–4.636, P<0.001) (Table 2).
 |
Table 2 Logistics Analysis of miRNAs and PMN-Related Membrane Proteins from Plasma-EVs in Sepsis-Induced Organ Dysfunction |
Enhanced analysis of plasma-EVs panel and their association with septic severity and 28-day mortality using logistic regression. The results showed that highly expressed of miR-34a-5p (OR=2.291, 95% CI 1.303–4.030, P=0.004), miR-4772-3p (OR=4.280, 95% CI 1.644–11.144, P=0.003), MMP9 (OR=1.422, 95% CI 1.060–1.907, P=0.019), and ITGAM (OR=0.996, 95% CI 0.992–0.999, P=0.025) independently predict the occurrence of septic shock. Further investigation revealed that an independent prognostic factor for sepsis 28-day overall survival included miR-4772-3p (OR=3.443, 95% CI 1.447–8.189, P=0.005), MMP9 (OR=1.536, 95% CI 1.138–2.074, P=0.005), and ITGAM (OR=1.004, 95% CI 1.000–1.008, P=0.042) (Table 3).
 |
Table 3 Logistics Analysis of miRNAs and PMN-Related Membrane Proteins from Plasma-EVs for Severity and Mortality in Sepsis |
EVs-Based Biomarkers Were Able to Predict Septic Organ Dysfunction and Prognosis
Six prominent indicators (miR-34a-5p, miR-503-5p, miR-4772-3p, ITGAM, MPO, and MMP9) identified in this study were used to evaluate their diagnostic performance. When used individually, MMP9, ITGAM, miR-34a-5p, and miR-4772-3p exhibited AUC values of 0.733, 0.670, 0.803 and 0.786, respectively, in distinguishing S-ARDS patients from Non-ARDS, and the AUC of multiple markers (0.911) showed higher sensitivity and specificity. In the S-ALF prediction, ROC analyses revealed that the AUC values of MPO and ITGAM were 0.887 and 0.786, respectively. When combined, these two membrane proteins significantly enhanced the discrimination efficiency of S-ALF, yielding an AUC value of 0.919. Furthermore, miR-4772-3p exhibited the best performance for the diagnosis of S-AKI, with an AUC value of 0.827. We combined miR-34a-5p and miR-503-5p with MMP9 to perform ROC analysis of septic shock, and found that this combination achieved a higher AUC (0.884) than miR-34a-5p, miR-503-5p, and MMP9 alone (0.77, 0.735, and 0.822, respectively). Notably, the AUC of multiple markers (0.870) was significantly greater than that of MMP9, ITGAM, or miR-4772-3p alone for 28-day mortality (Figure 6A). Next, the prognosis of miRNAs and PMN-related membrane proteins from plasma-EVs in sepsis was analyzed. Kaplan-Meier survival analysis showed that individuals with high expression levels of miR-34a-5p, miR-4772-3p, ITGAM, and MMP9 had worse prognosis than those with low expression levels (Figure 6B). Collectively, the levels of miR-34a-5p, miR-4772-3p, ITGAM, and MMP9 could be used as potential prognostic predictors in patients with sepsis.
Figure 6 Continued.
We constructed a target interaction regulatory network diagram to depict the functions of DE-miRNAs and PMN-related membrane proteins in plasma-EVs. The ITGAM, MPO, and MMP9 levels were highly correlated with miR-34a-5p expression. The key pathways included the Hedgehog signaling pathway, Rap1 signaling pathway, Hippo signaling pathway, ErbB signaling pathway, Ras signaling pathway, MAPK signaling pathway, PI3K-Akt signaling pathway, NF-κB signaling pathway, TNF signaling pathway, and autophagy (Figure 7).
 |
Figure 7 miRNAs and PMN-related membrane proteins from plasma-EVs with differential expression target the interaction regulatory network diagram. |
Plasma-EVs Suppressed PMNs Autophagy
Enrichment pathway analysis revealed that the target genes are mainly involved in several inflammatory and autophagy-related pathways. However, the role of plasma-EVs in the regulation of PMNs autophagy during sepsis remains unclear. We hypothesized that plasma-EVs contain PMN-EVs and play a pivotal role in the response to damage in target cells during sepsis. Immunoblotting analysis further confirmed that the PMN-specific markers CXCR2, CXCR4, MMP9, and ITGAM were expressed in the plasma-EVs of healthy controls and patients with sepsis (Figure 8A). We co-incubated plasma-EVs with PMNs directly. Intriguingly, we observed that DiR-labelled EVs were taken up by PMNs in vitro (Figure 8B). Immunoblotting analysis demonstrated that the expression of ATG5, ATG7, Beclin-1, and LC3B decreased after stimulation of PMNs with plasma-EVs from sepsis patients, whereas the level of P62 was increased (Figure 8C). An experimental sepsis model was induced in rats by intraperitoneal injection of 10 mg/kg LPS. Twenty-four hours after the administration of LPS, significant differences were observed in serum tests between the Saline group and the LPS group, as indicated by several indices (TNF-α, IL-1β, and IL-6) (Figure S3). Together, these data suggest that a sepsis model was successfully established in rats. To further verify the role of plasma-EVs in the induction of PMNs autophagy after LPS induction, GW4869, an EV-release inhibitor, was used to reduce EV secretion in the systemic circulation. GW4869 treatment significantly increased the levels of ATG5, ATG7, Beclin-1, and LC3B in rats with sepsis while reducing the level of P62 (Figure 8D). Furthermore, the administration of LPS-EVs to healthy rats also significantly suppressed PMNs autophagy in vivo, whereas Saline-EVs had no such effect, once again demonstrating the close association between septic plasma-EVs and PMNs autophagy. Moreover, we further validated these findings in vivo, treatment with enoxaparin, an inhibitor of EV uptake, significantly stabilized PMNs autophagy as evidenced by increased levels of ATG5, ATG7, Beclin-1 and LC3B and decreased expression of P62 compared to LPS-EVs (Figure 8E). These observations suggest that plasma-EVs from sepsis patients suppress PMNs autophagy both in vivo and in vitro.
Figure 8 Continued.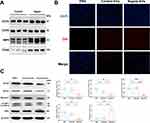
Discussion
The present study suggests that miRNAs and PMN-related membrane proteins from plasma-EVs have the potential to serve as biomarkers for sepsis-induced organ dysfunction and prognosis. More specifically, the high expression of these factors (miR-34a-5p, miR-503-5p, miR-4772-3p, ITGAM, MPO, and MMP9) in plasma-EVs is associated with septic organ dysfunction. Notably, we observed a significant correlation between elevated levels of miR-34a-5p, miR-4772-3p, ITGAM, and MMP9 with unfavorable survival outcomes. Furthermore, PMN-EVs were released into the plasma, and sepsis-induced plasma-EVs suppressed PMNs autophagy, which could be reversed by EV inhibitors such as GW4869 and enoxaparin.
PMNs, which are professional killer cells of the innate immune system, have gained recognition.15 The potent antimicrobial effect of PMNs confers them with a dual role as both crucial guardians of host defenses and detrimental facilitators of tissue damage in an uncontrolled inflammatory state.16 However, little attention has been paid to the changes in the PMN-EVs within the body, despite substantial evidence suggesting that PMN-EVs are pivotal components of their in vivo functionality.17–19 However, the role of PMN-related membrane proteins in sepsis remains unclear. Several proteins enriched in plasma-EVs identified in this study have been previously implicated in disease progression or have been identified as risk factors for sepsis. EV-derived ITGAM mediates severe acute pancreatitis-related acute lung injury.20 Moreover, elevated plasma levels of MPO-DNA complexes have been found to be associated with a poor outcome in acute liver failure, suggesting that NET formation contributes to the disease progression.21 Another sepsis-predictive EV protein, MMP9, identified in our study, has been shown to be associated with septic shock and significantly correlated with the SOFA score.22 Herein, we report that as sepsis progresses, PMN-EVs can be released into the bloodstream, partly participating in plasma-EVs, suggesting that shifting the focus from PMNs to their EV-related membrane proteins might offer an innovative diagnostic strategy.
EVs, as intricate mediators of sepsis, play pleiotropic roles in both alleviating and exacerbating the disease’s progression.23 Recent studies have pointed out that EVs mediate intracellular communication by delivering functional proteins, mRNA transcription products, and miRNAs from maternal cells to recipient cells.24 Most notably, miRNAs are emerging as key players in the pathogenesis of sepsis and have been implicated in the regulation of cell death, cell metabolism, and inflammatory.25–27 The growing therapeutic potential of specific EV-derived miRNAs, such as miR-125b-5p, miR-193b-5p, miR-377-3p, and miR-709, has been identified in the treatment of sepsis.28–31 Several miRNAs enriched in plasma-EVs, identified in this study, have previously been implicated in disease development or identified as risk factors for sepsis. miR-34a-5p expression was significantly higher in plasma-EVs from septic mice than in those from the sham group, and it has been found to accelerate sepsis-induced apoptosis and exacerbate lung injury, suggesting its involvement in the pathogenesis of sepsis.32,33 miR-503-5p is recognized as a signature molecule in plasma-EVs of severe acute pancreatitis-associated lung injury and is an important regulator in the rat ALI model.34,35 Serum exosomal miR-4772-3p has previously been identified as a prognostic marker for tumor recurrence in stage II and III colon cancer;36 however, the precise role of miR-4772-3p in sepsis remains unclear. If patients are likely to have poor outcomes at the onset of sepsis, the early administration of tailored therapy may improve their outcomes. Our findings further support the notion that plasma-EV miRNAs could potentially serve as diagnostic biomarkers for septic organ dysfunction and prognosis.
EV-mediated intercellular crosstalk plays a critical role in sepsis-induced organ dysfunction.23 Specific immune cells release EVs loaded with pro-inflammatory signals, thereby exacerbating inflammation and leading to further organ dysfunction. Current evidence suggests that ARDS, ALF, and AKI are common complications of sepsis, and are associated with increased morbidity and mortality.37 Previous studies have shown that PMN-EVs, abundant in MMP9, undermine cellular cohesion and disrupt tissue structure integrity, resulting in extensive tissue damage and impairment of epithelial barrier function.38 Additionally, miR-30d-5p in PMN-EVs induces M1 macrophage polarization and triggers macrophage pyroptosis in ALI, while modulation of the cross-talk between PMNs and macrophages attenuates lung injury.19 Furthermore, LPS induced macrophage-EVs act on hepatocytes, activating NLRP3 inflammasome and caspase-1, subsequently leading to liver injury.39 Moreover, exosomal miR-19b-3p mediated the cross-talk between tubular epithelial cells and macrophages and contributed to M1 macrophage activation.27
Plasma-EVs exhibit bidirectional regulatory effects on disease progression. Plasma-EVs from acute pancreatitis mice contribute to pancreatitis-associated lung injury by triggering NLRP3-dependent pyroptosis in alveolar macrophages.40 Conversely, plasma-EVs from young healthy humans improve functional recovery by counteracting ferroptotic injury via regulation of the P53/SLC7A11/GPX4 axis after Intracerebral hemorrhage.41 In addition, healthy plasma-EVs circ_0001785, for atherogenesis, which can reduce endothelial cell injury and thus delay atherogenesis through the miR-513a-5p/TGFBR3 ceRNA network mechanism.42 Therefore, targeting plasma-EVs may be a promising avenue for the treatment of sepsis.
Autophagy is a self-destructive process that is crucial for the removal of damaged organelles, disordered proteins, and intracellular infections.43,44 Emerging evidence has shown that enhanced autophagy may contribute to a protective effect on organs during sepsis, ameliorating inflammation, lung injury, kidney injury, and cardiac injury.45–49 In contrast, a few studies have demonstrated the opposite. The increase in plasma mtDNA and protein levels during sepsis suggests that promoting autophagy may trigger an inflammatory response in the immune cells.50 Although drug interventions targeting PMNs death in sepsis remain to be investigated, there is evidence that heparin neutralizes extracellular histones and reduces their toxicity, thereby increasing survival in animal models of sepsis.51,52 We found that plasma-EVs from patients with sepsis suppressed autophagy in PMNs. Additionally, we found that GW4869 and enoxaparin significantly stabilized sepsis-induced PMNs autophagy, as numerous studies have shown that targeting PMNs cell death may be a promising treatment for sepsis.
Our study had several limitations that warrant consideration. First, the decision to utilize intraperitoneal injection of LPS for the establishment of a sepsis model in this study was made based on the following considerations. There is no question that cecal ligation and puncture (CLP) remains a classic model of sepsis. However, the CLP model presents challenges in controlling the amount of bowel leakage, resulting in a wide range of variation in sepsis outcomes. Additionally, when assessing the impact on acute inflammation, the LPS model is the most suitable since LPS treatment elicits a stronger response, as systemic effects are easily identifiable, measurable, and reproducible.53 A more comprehensive comparison of the effects of LPS and CLP models warrants further studies in the future. Second, the investigation of molecular mechanisms is subject to short-lived PMNs; therefore, further exploration will be performed. Third, the current study may be influenced by the high heterogeneity of EVs and frequent occurrence of multiple organ dysfunction in sepsis. Therefore, it is imperative to conduct a larger multicenter study to establish a robust correlation between miRNAs and PMN-related membrane proteins, and their long-term prognostic implications in sepsis. Finally, although our study highlighted the role of miRNAs and PMN-related membrane proteins in plasma-EVs in sepsis, further in-depth research is required to investigate the contributions of other components, such as proteins, DNA, and mRNA, to develop new therapeutic strategies for managing human sepsis.
Conclusions
Our study successfully identified a panel of EV-based biomarkers for diagnosing septic organ dysfunction and evaluating the prognosis. This will facilitate a more comprehensive investigation into the pathogenesis of sepsis, discovery of biomarkers for predicting disease progression, and development of EV-based therapeutic interventions (Figure 9).
Abbreviations
ARDS, acute respiratory distress syndrome; ALI, acute lung injury; AKI, acute kidney injury; APACHE II, acute physiology and chronic health evaluation; Ct, cycle threshold; CLP, cecal ligation and puncture; EV, extracellular vesicles; ELISA, enzyme-linked immunosorbent assay; GO, Gene ontology; IL-1β, interleukin-1β; IL-6, interleukin-6; KEGG, kyoto encyclopedia of genes and genomes; LPS, lipopolysaccharide; miRNA, microRNA; PMNs, polymorphonuclear neutrophils; PCI, peritoneal contamination and infection; RT-qPCR, quantitative real-time polymerase chain reaction; SOFA, sequential organ failure assessment; SEC, molecular size exclusion; TNF-α, tumor necrosis factor-α; TEM, transmission electron microscopy; UC, ultracentrifugation.
Data Sharing Statement
All data generated or analyzed during the current study are included in this published article (and its supplementary material files).
Ethical Approval
All the protocols, procedures, and subject/patient recruitment procedures described in this study were approved by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University, China (No. 2023-K128-01). This study was conducted in accordance with the principles of the Declaration of Helsinki. Written informed consent was obtained from all the subjects and patients. All animal experiments were conducted in accordance with the National Institutes of Health “Guidelines for Care and Use of the Laboratory Animals” and approved by the Animal Ethics Committee of Guangxi Medical University (Approval No. 202305565). Every effort was made to minimize the number of rats used and their suffering.
Acknowledgments
We are grateful to all participants and their families, hospital staff, and our lab members for their invaluable assistance throughout the study. We acknowledge Figdraw (www.figdraw.com) for assistance in creating Figure 9 for this study.
Funding
This work was supported by the National Natural Science Foundation of China (No. 82360024) and Innovation Project of Guangxi Graduate Education (No. YCBZ2022096, YCSW2023222, YCSW2024249).
Disclosure
No potential conflict of interest was reported by the author(s).
References
1. Singer M, Deutschman CS, Seymour CW, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA. 2016;315(8):801–810. doi:10.1001/jama.2016.0287
2. Marshall JC. Why have clinical trials in sepsis failed? Trends Mol Med. 2014;20(4):195–203. doi:10.1016/j.molmed.2014.01.007
3. Stanski NL, Wong HR. Prognostic and predictive enrichment in sepsis. Nat Rev Nephrol. 2020;16(1):20–31. doi:10.1038/s41581-019-0199-3
4. Seymour CW, Kennedy JN, Wang S, et al. Derivation, validation, and potential treatment implications of novel clinical phenotypes for sepsis. JAMA. 2019;321(20):2003–2017. doi:10.1001/jama.2019.5791
5. DeMerle KM, Angus DC, Baillie JK, et al. Sepsis subclasses: a framework for development and interpretation. Crit Care Med. 2021;49(5):748–759. doi:10.1097/ccm.0000000000004842
6. Pickkers P, Kox M. Towards precision medicine for sepsis patients. Crit Care. 2017;21(1):11. doi:10.1186/s13054-016-1583-z
7. Lim CZJ, Zhang Y, Chen Y, et al. Subtyping of circulating exosome-bound amyloid β reflects brain plaque deposition. Nat Commun. 2019;10(1):1144. doi:10.1038/s41467-019-09030-2
8. Raeven P, Zipperle J, Drechsler S. Extracellular vesicles as markers and mediators in sepsis. Theranostics. 2018;8(12):3348–3365. doi:10.7150/thno.23453
9. Li L, Zhang Q, Feng Y, et al. A Novel Serum Exosomal miRNA Signature in the Early Prediction of Persistent Organ Failure in Patients with Acute Pancreatitis. Ann Surg. 2024. doi:10.1097/sla.0000000000006229
10. Gong T, Zhang X, Peng Z, et al. Macrophage-derived exosomal aminopeptidase N aggravates sepsis-induced acute lung injury by regulating necroptosis of lung epithelial cell. Commun Biol. 2022;5(1):543. doi:10.1038/s42003-022-03481-y
11. Park C, Lei Z, Li Y, et al. Extracellular vesicles in sepsis plasma mediate neuronal inflammation in the brain through miRNAs and innate immune signaling. J Neuroinflamm. 2024;21(1):252. doi:10.1186/s12974-024-03250-0
12. Qi X, Yu Y, Sun R, et al. Identification and characterization of neutrophil heterogeneity in sepsis. Crit Care. 2021;25(1):50. doi:10.1186/s13054-021-03481-0
13. Jacobi J. The pathophysiology of sepsis - 2021 update: part 2, organ dysfunction and assessment. Am J Health Syst Pharm. 2022;79(6):424–436. doi:10.1093/ajhp/zxab393
14. Ye R, Lin Q, Xiao W, et al. miR-150-5p in neutrophil-derived extracellular vesicles associated with sepsis-induced cardiomyopathy in septic patients. Cell Death Discovery. 2023;9(1):19. doi:10.1038/s41420-023-01328-x
15. Ley K, Hoffman HM, Kubes P, et al. Neutrophils: new insights and open questions. Sci Immunol. 2018;3(30). doi:10.1126/sciimmunol.aat4579
16. Liew PX, Kubes P. The neutrophil’s role during health and disease. Physiol Rev. 2019;99(2):1223–1248. doi:10.1152/physrev.00012.2018
17. Marki A, Buscher K, Lorenzini C, et al. Elongated neutrophil-derived structures are blood-borne microparticles formed by rolling neutrophils during sepsis. J Exp Med. 2021;218(3). doi:10.1084/jem.20200551
18. Johnson BL 3rd, Midura EF, Prakash PS, et al. Neutrophil derived microparticles increase mortality and the counter-inflammatory response in a murine model of sepsis. Biochim Biophys Acta Mol Basis Dis. 2017;1863(10 Pt B):2554–2563. doi:10.1016/j.bbadis.2017.01.012
19. Jiao Y, Zhang T, Zhang C, et al. Exosomal miR-30d-5p of neutrophils induces M1 macrophage polarization and primes macrophage pyroptosis in sepsis-related acute lung injury. Crit care. 2021;25(1):356. doi:10.1186/s13054-021-03775-3
20. Hu Q, Zhang S, Yang Y, et al. Extracellular vesicle ITGAM and ITGB2 mediate severe acute pancreatitis-related acute lung injury. ACS Nano. 2023;17(8):7562–7575. doi:10.1021/acsnano.2c12722
21. von Meijenfeldt FA, Stravitz RT, Zhang J, et al. Generation of neutrophil extracellular traps in patients with acute liver failure is associated with poor outcome. Hepatology. 2022;75(3):623–633. doi:10.1002/hep.32174
22. Hong Y, Chen L, Sun J, et al. Single-cell transcriptome profiling reveals heterogeneous neutrophils with prognostic values in sepsis. iScience. 2022;25(11):105301. doi:10.1016/j.isci.2022.105301
23. Gong T, Liu YT, Fan J. Exosomal mediators in sepsis and inflammatory organ injury: unraveling the role of exosomes in intercellular crosstalk and organ dysfunction. Mil Med Res. 2024;11(1):24. doi:10.1186/s40779-024-00527-6
24. Sohail AM, Khawar MB, Afzal A, Hassan A, Shahzaman S, Ali A. Multifaceted roles of extracellular RNAs in different diseases. Mil Med Res. 2022;9(1):43. doi:10.1186/s40779-022-00405-z
25. Zhang L, Meng H, Geng Q. Commentary: peripheral Circulating Exosome-Mediated Delivery of miR-155 as a Novel Mechanism for Acute Lung Inflammation. Front Med. 2020;7:507. doi:10.3389/fmed.2020.00507
26. Liu F, Peng W, Chen J, et al. Exosomes derived from alveolar epithelial cells promote alveolar macrophage activation mediated by miR-92a-3p in sepsis-induced acute lung injury. Front Cell Infect Microbiol. 2021;11:646546. doi:10.3389/fcimb.2021.646546
27. Lv LL, Feng Y, Wu M, et al. Exosomal miRNA-19b-3p of tubular epithelial cells promotes M1 macrophage activation in kidney injury. Cell Death Differ. 2020;27(1):210–226. doi:10.1038/s41418-019-0349-y
28. Shen K, Wang X, Wang Y, et al. miR-125b-5p in adipose derived stem cells exosome alleviates pulmonary microvascular endothelial cells ferroptosis via Keap1/Nrf2/GPX4 in sepsis lung injury. Redox Biol. 2023;62:102655. doi:10.1016/j.redox.2023.102655
29. Dos Santos CC, Amatullah H, Vaswani CM, et al. Mesenchymal stromal (stem) cell therapy modulates miR-193b-5p expression to attenuate sepsis-induced acute lung injury. Eur Respir J. 2022;59:2004216. doi:10.1183/13993003.54216-2020
30. Wei X, Yi X, Lv H, et al. Correction: microRNA-377-3p released by mesenchymal stem cell exosomes ameliorates lipopolysaccharide-induced acute lung injury by targeting RPTOR to induce autophagy. Cell Death Dis. 2020;11(9):746. doi:10.1038/s41419-020-02976-y
31. Yang J, Huang X, Yu Q, et al. Extracellular vesicles derived from M2-like macrophages alleviate acute lung injury in a miR-709-mediated manner. J Extracell Vesicles. 2024;13(4):e12437. doi:10.1002/jev2.12437
32. Xu J, Feng Y, Jeyaram A, Jay SM, Zou L, Chao W. Circulating plasma extracellular vesicles from septic mice induce inflammation via MicroRNA- and TLR7-dependent mechanisms. J Immunol. 2018;201(11):3392–3400. doi:10.4049/jimmunol.1801008
33. Khan MJ, Singh P, Jha P, et al. Investigating the link between miR-34a-5p and TLR6 signaling in sepsis-induced ARDS. 3 Biotech. 2023;13(8):282. doi:10.1007/s13205-023-03700-1
34. Xiong Y, Chen X, Yang X, et al. miRNA transcriptomics analysis shows miR-483-5p and miR-503-5p targeted miRNA in extracellular vesicles from severe acute pancreatitis-associated lung injury patients. Int Immunopharmacol. 2023;125(Pt A):111075. doi:10.1016/j.intimp.2023.111075
35. Lee W, Kim I, Shin S, et al. Expression profiling of microRNAs in lipopolysaccharide-induced acute lung injury after hypothermia treatment. Mol Cell Toxicol. 2016;12(3):243–253. doi:10.1007/s13273-016-0029-7
36. Liu C, Eng C, Shen J, et al. Serum exosomal miR-4772-3p is a predictor of tumor recurrence in stage II and III colon cancer. Oncotarget. 2016;7(46):76250–76260. doi:10.18632/oncotarget.12841
37. Kumar S, Payal N, Srivastava VK, Kaushik S, Saxena J, Jyoti A. Neutrophil extracellular traps and organ dysfunction in sepsis. Clin Chim Acta. 2021;523:152–162. doi:10.1016/j.cca.2021.09.012
38. Butin-Israeli V, Houser MC, Feng M, et al. Deposition of microparticles by neutrophils onto inflamed epithelium: a new mechanism to disrupt epithelial intercellular adhesions and promote transepithelial migration. FASEB J. 2016;30(12):4007–4020. doi:10.1096/fj.201600734R
39. Wang G, Jin S, Ling X, et al. Proteomic profiling of lps-induced macrophage-derived exosomes indicates their involvement in acute liver injury. Proteomics. 2019;19(3):e1800274. doi:10.1002/pmic.201800274
40. Wu XB, Sun HY, Luo ZL, Cheng L, Duan XM, Ren JD. Plasma-derived exosomes contribute to pancreatitis-associated lung injury by triggering NLRP3-dependent pyroptosis in alveolar macrophages. Biochim Biophys Acta Mol Basis Dis. 2020;1866(5):165685. doi:10.1016/j.bbadis.2020.165685
41. Yang W, Ding N, Luo R, et al. Exosomes from young healthy human plasma promote functional recovery from intracerebral hemorrhage via counteracting ferroptotic injury. Bioact Mater. 2023;27:1–14. doi:10.1016/j.bioactmat.2023.03.007
42. Tong X, Dang X, Liu D, et al. Exosome-derived circ_0001785 delays atherogenesis through the ceRNA network mechanism of miR-513a-5p/TGFBR3. J Nanobiotechnol. 2023;21(1):362. doi:10.1186/s12951-023-02076-x
43. Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221(1):3–12. doi:10.1002/path.2697
44. Bhattacharya A, Wei Q, Shin JN, et al. Autophagy is required for neutrophil-mediated inflammation. Cell Rep. 2015;12(11):1731–1739. doi:10.1016/j.celrep.2015.08.019
45. Jia J, Gong X, Zhao Y, et al. Autophagy enhancing contributes to the organ protective effect of alpha-lipoic acid in septic rats. Front Immunol. 2019;10:1491. doi:10.3389/fimmu.2019.01491
46. Zhao H, Chen H, Xiaoyin M, et al. Autophagy activation improves lung injury and inflammation in sepsis. Inflammation. 2019;42(2):426–439. doi:10.1007/s10753-018-00952-5
47. Zuo Y, Dang R, Peng H, Wu Z, Yang Y. LL-37 exacerbates local inflammation in sepsis-induced acute lung injury by preventing mitochondrial DNA (mtDNA) degradation-induced autophagy. Med Sci Monit. 2019;25:6193–9203. doi:10.12659/msm.915298
48. Zhao Y, Feng X, Li B, et al. Dexmedetomidine protects against lipopolysaccharide-induced acute kidney injury by enhancing autophagy through inhibition of the PI3K/AKT/mTOR pathway. Front Pharmacol. 2020;11:128. doi:10.3389/fphar.2020.00128
49. Hsieh CH, Pai PY, Hsueh HW, Yuan SS, Hsieh YC. Complete induction of autophagy is essential for cardioprotection in sepsis. Ann Surg. 2011;253(6):1190–1200. doi:10.1097/SLA.0b013e318214b67e
50. Unuma K, Aki T, Funakoshi T, Hashimoto K, Uemura K. Extrusion of mitochondrial contents from lipopolysaccharide-stimulated cells: involvement of autophagy. Autophagy. 2015;11(9):1520–1536. doi:10.1080/15548627.2015.1063765
51. Cheng Z, Abrams ST, Alhamdi Y, et al. Circulating histones are major mediators of multiple organ dysfunction syndrome in acute critical illnesses. Crit Care Med. 2019;47(8):e677–e684. doi:10.1097/ccm.0000000000003839
52. Wildhagen KC, García de Frutos P, Reutelingsperger CP, et al. Nonanticoagulant heparin prevents histone-mediated cytotoxicity in vitro and improves survival in sepsis. Blood. 2014;123(7):1098–1101. doi:10.1182/blood-2013-07-514984
53. Seemann S, Zohles F, Lupp A. Comprehensive comparison of three different animal models for systemic inflammation. J Biomed Sci. 2017;24(1):60. doi:10.1186/s12929-017-0370-8
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.









