Back to Journals » Journal of Inflammation Research » Volume 18
Multi-Target Synergy Against Acute Lung Injury: Systems Pharmacology Decoding Dahuang-Huangqin Herb Pair’s Therapeutic Mechanism
Authors Zhang J, Zhu X, Chen X, Liu H, Yan Z, Chen Z, Li Q, Yu W
Received 8 April 2025
Accepted for publication 5 July 2025
Published 18 July 2025 Volume 2025:18 Pages 9413—9441
DOI https://doi.org/10.2147/JIR.S527378
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Jinquan Zhang,1– 4 Xuejiao Zhu,5 Xiaona Chen,2 Haichao Liu,1 Zhengzheng Yan,6 Zhixia Chen,2 Quan Li,1,2,4 Weifeng Yu3
1Guangzhou University of Chinese Medicine, Guangzhou, 510006, People’s Republic of China; 2Department of Anesthesiology, National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital & Shenzhen Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Shenzhen, 518116, People’s Republic of China; 3Department of Anesthesiology, Renji Hospital, Shanghai Jiao Tong University School of Medicine, Key Laboratory of Anesthesiology (Shanghai Jiao Tong University), Ministry of Education, Shanghai, People’s Republic of China; 4Shenzhen Clinical Medical College, Guangzhou University of Chinese Medicine, Shenzhen, 518100, People’s Republic of China; 5Department of Anesthesiology, The Second Affiliated Hospital of Soochow University, Suzhou, Jiangsu, 215004, People’s Republic of China; 6Department of Anesthesiology, Dongguan Key Laboratory of Anesthesia and Organ Protection, the Tenth Affiliated Hospital of Southern Medical University, Dongguan, Guangdong, People’s Republic of China
Correspondence: Zhixia Chen, Email [email protected] Quan Li, Email [email protected]
Aim: This study investigates the therapeutic mechanisms of Radix et Rhizoma Rhei-Scutellaria baicalensis Georgi (DH-HQ) herb pair against acute lung injury (ALI) using integrative network pharmacology and multi-omics approaches.
Methods: We combined bioactive compound screening of DH-HQ with ALI-related transcriptomics to construct dual interaction networks (herb-compound-target and protein-protein interactions). Core pathways were identified via bioinformatic analyses (GO/KEGG/GSEA) and validated through Mendelian randomization (to confirm gene causality), single-cell RNA sequencing (cell-type specificity), molecular docking (binding stability assessment), and in vivo ALI models (pathological/clinical validation).
Results: Our analysis identified 207 active DH-HQ components and 93 therapeutic targets associated with ALI. PPI network analysis revealed five key regulatory targets: GAPDH, IL1B, IL10, MMP9, and TP53. Functional enrichment analyses demonstrated DH-HQ’s modulation of critical pathways including HIF-1 signaling and cytokine-cytokine receptor interaction. Molecular docking revealed strong compound-target binding affinities (energies: − 8.8 to − 7.3 kcal/mol), with emodin and baicalin showing the highest stability. In vivo, DH-HQ co-treatment significantly attenuated LPS-induced ALI, evidenced by reduced clinical scores and body weight recovery (p< 0.05). Multi-modal validation prioritized LTB4R/MMP9/POLR1B as diagnostic hubs (AUC > 0.70).
Conclusion: This study elucidates DH-HQ’s multi-target mechanism in mitigating ALI via synergistic bioactive components (emodin and baicalin) and key pathway modulation. Preclinical evidence confirms efficacy through diagnostic hub genes and compound interactions, yet clinical application demands pharmacodynamic refinement.
Keywords: acute lung injury (ALI), radix et Rhizoma Rhei (DaHuang), Scutellaria baicalensis Georgi (HuangQin), network pharmacology, single-cell sequencing, Mendelian randomization
Introduction
Acute lung injury (ALI) is a respiratory condition induced by a variety of pulmonary and extrapulmonary factors, characterized by increased alveolar permeability, refractory hypoxemia, and pulmonary edema. The progression of ALI might result in acute respiratory distress syndrome (ARDS), which is a distinct phase of the same disease process. ARDS is associated with increased lung tissue damage and a higher mortality rate.1 Although treatment procedures have advanced, the fatality rate of ALI remains high. According to a clinical observational study involving 145 pediatric intensive care units across 27 countries from 2016 to 2017, the incidence and mortality rates of ALI in children were found to be 17% and 3.2%, respectively.2 Additionally, the mortality rate from ALI rises with age. The mortality rate associated with ALI is 24% among individuals aged 15–19 years and surges to 60% among patients aged ≥80 years.3,4 Therefore, it is crucial to develop novel and efficacious medications for the treatment of ALI.5
Radix et Rhizoma Rhei (known as DaHuang in Chinese, DH) is a renowned herbal remedy in traditional Chinese medicine (TCM) that is frequently employed in the treatment of hypertension, atherosclerosis, gastritis, and hepatitis.6 According to the principles of TCM, DH is believed to have the effects of activating blood, dissipating stasis, and removing toxicity.7 For about 2000 years, Scutellaria baicalensis Georgi (known as Huangqin in Chinese, HQ) has been an extensively utilized medicinal material in China. It has received official recognition through its inclusion in the Chinese Pharmacopoeia, British Pharmacopoeia (BP 2018), and European Pharmacopoeia (EP 9.0).8,9 HQ has extensive historical usage in the treatment of respiratory, gastrointestinal, and inflammatory ailments. Fifty-six flavonoid compounds, including wogonoside, baicalein, baicalin, and wogonin, have been extracted from HQ.10 The DH-HQ herb pair is among the top ten herb pairs in TCM formulas used to treat ALI. Additionally, DH and HQ are placed first and third, respectively, on the list of high-frequency Chinese medicines. Both substances possess the properties of heat-clearing and detoxification, making them commonly employed in conjunction for the treatment of patients with ALI who manifest symptoms of intestinal heat, solid viscera, and heat toxin stagnation syndrome.11,12 Prior studies have focused on individual effects of DH or HQ in ALI treatment, but the synergistic mechanisms of this herb pair remain unclear, particularly how their bioactive components collectively modulate key pathophysiological processes like inflammation, oxidative stress, and gut barrier function.
Network pharmacology is a pragmatic method that utilizes bioinformatics and systems biology to examine the molecular relationships between medications and their therapeutic targets.13 This approach has become instrumental in decoding the holistic mechanisms of traditional Chinese medicine (TCM).14,15 Complementarily, molecular docking simulations elucidate ligand-receptor binding affinity, prioritizing bioactive compounds with optimal target engagement potential.16,17 These methodologies synergistically bridge TCM’s multi-target nature with modern translational research paradigms.
We hypothesize that the DH-HQ herb pair alleviates ALI through emodin-baicalein-mediated multi-target regulation of inflammatory pathways. Using network pharmacology, we first identified core bioactive compounds and ALI-related targets, then validated potential interactions through molecular docking analysis. Further investigation of the therapeutic mechanism was conducted through pathway enrichment analysis and in vivo experimental validation. By integrating computational predictions with experimental verification, this study systematically established a comprehensive herb-target-pathway network that elucidates the multi-target pharmacological mechanisms responsible for the observed anti-inflammatory effects. A flowchart summarizing the study design is presented in Figure 1.
 |
Figure 1 A schematic diagram of the research process. |
Materials and Methods
Collection of Active Ingredients and Potential Targets
Screening of the Active Ingredients in DH-HQ
We employed the following databases to ascertain the constituent elements of DH-HQ, which are enumerated below: the TCMSP database18 (A database of systems pharmacology for drug discovery from herbal medicines. https://old.tcmsp-e.com/tcmsp.php, Ver.2.3), the BATMAN-TCM database19 (An online bioinformatics analysis tool specially designed for research on molecular mechanism of TCM. http://bionet.ncpsb.org.cn/batman-tcm/index.php, ver2.0), the ETCM database20 (A systematic and normalized database of the prevalent herbal drugs and formulas in TCM, as well as their ingredients. http://www.tcmip.cn/ETCM/index.php/Home/Index/).
Prediction of the Targets of DH-HQ
Within the TCMSP database, we utilized a drug-likeness (DL) threshold of ≥0.18 and an oral bioavailability (OB) threshold of ≥30% to selectively screen the relevant constituents. DL refers to the resemblance or existence of certain functional groups in the chemical composition of compounds to those found in drugs approved by the Food and Drug Administration (FDA). This similarity aids in enhancing the pharmacokinetics and properties of the drug, such as solubility and chemical stability. OB is a crucial pharmacokinetic property of oral drugs. It is defined by the FDA as the speed at which the active component is absorbed and the amount that is absorbed from the drug and becomes accessible at the intended site of action. By following the aforementioned procedure, we acquired the SMILES codes for each compound from PubChem21 (https://pubchem.ncbi.nlm.nih.gov/) and forecasted the targets of the active constituents using Swiss Target Prediction22 (http://www.swisstargetprediction.ch/). SMILES codes serve as a textual representation of the molecular structures of compounds. The targets that were evaluated on the Swiss Target Prediction website were filtered according to the “Probability > 0” criterion.
Similarly, we applied “Score cutoff > 20, Adjusted P-value > 0.05” and “Reliability score > 0.8” as the filtering criteria for the BATMAN-TCM database and the ETCM database, respectively, to filter the active ingredients corresponding to the targets. Ultimately, we employed the UniProt database23 (https://www.uniprot.org) to standardize the target UniProt ID into target gene names. Subsequently, we then obtained the desired drug targets by removing duplicate compounds from the ones screened from the above databases. The selected databases provide complementary analytical strengths: BATMAN-TCM, a pioneering bioinformatics platform specialized in TCM molecular mechanism exploration, employs structural similarity-based target prediction; ETCM integrates standardized herb/formula data with chemical fingerprint-derived target insights for enhanced reliability; while TCMSP prioritizes pharmacokinetically viable candidates through DL/OB criteria. This multi-source strategy mitigates systematic biases inherent to single-database approaches.24
Identification of Potential Targets in ALI
Access a wealth of information about gene sequences, expression, structure, variation, function, interactions, and more from a wide variety of taxa through NCBI-gene (https://www.ncbi.nlm.nih.gov/gene/), a public library of gene-centric data. The Online Mendelian Inheritance in Man25 (OMIM, https://omim.org/, Updated: Nov 3, 2023) database contains over 16,000 genes and information on every known Mendelian condition. It is an authoritative and comprehensive collection of human genes and genetic traits. A comprehensive collection of annotated and predicted human genes is available in GeneCards26 (https://www.genecards.org/, Version 5.18, Updated: Oct 5, 2023), which is a searchable merged database. A discovery platform known as DisGeNET27 (https://www.disgenet.org/, Version 7) comprises genes and variants that have been associated with human diseases.
We conducted a search across four databases using the keywords “acute lung injury” and “acute respiratory distress syndrome” specifically for the “Organism: Homo sapiens”. As a result, we found the gene targets associated with both disorders.
Screening of ARDS-Related Genes Through the GEO Database
Leveraging peripheral blood samples derived from Homo sapiens, we accessed gene expression profiles associated with ARDS (GSE243066)28 from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). The GSE243066 dataset, based on the GPL30209 sequencing platform, includes transcriptomic data from blood samples collected within 24 hours of hospital admission from 34 patients with ARDS and 15 healthy controls. This dataset provides a comprehensive integration of mRNA and miRNA expression analyses. In our investigation, we concentrated exclusively on the mRNA data, cross-referencing it with previously identified drug-disease intersection genes to pinpoint therapeutic targets that could play a crucial role in mediating drug efficacy under pathological conditions. For this dataset, we constructed a gene expression heatmap along with box plots to illustrate the differential expression levels.
Therapeutic Targets of DH-HQ for Treating ALI
To perform graphical analyses, we used two online Venn diagram tools: Venny 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/index.html) and jvenn29 (http://www.bioinformatics.com.cn/static/others/jvenn/example.html). We obtained the medication, illness, and their common targets from the databases. Both jvenn and Venny 2.1 make it easy to see how several sets overlap and vary.
Analysis and Establishment of PPI Networks
PPI is a visual representation technique used to illustrate the links and interactions between proteins. The intersection targets of DH-HQ and ALI were imported into the STRING database30 (https://string-db.org/,version 12.0), with the minimum required interaction score set to “highest confidence: 0.9” and the organism specified as “Homo sapiens”. This allowed us to obtain the PPI network relationship diagram and data, which were saved in a tab-separated values (tsv) format file. The data from the generated tsv format file was imported into Cytoscape 3.10.1 to visualize and create a PPI network. By utilizing the CytoNCA plugin in Cytoscape 3.10.1, we successfully derived the degree, eigenvector (EC), closeness centrality (CC), betweenness centrality, and other relevant characteristics of the intersection targets.
GO and KEGG Pathway Enrichment
To investigate the functional characteristics of DH-HQ in the treatment of ALI, we applied the DAVID database31,32 (https://david.ncifcrf.gov/) to explore the probable processes that underlie the primary targets. We specified the species as “Homo sapiens” and selected the annotation findings with a significance level of P <0.05 and FDR <0.05. We examined the GO functions, comprising the biological process (BP), cell component (CC), molecular function, and KEGG pathway. Bar charts, bubble charts, chord diagrams, and enrichment circle diagrams were used to display the top 10 entries from the GO analysis and the top 30 entries from the KEGG pathway analysis. We ranked the enriched genes from high to low and used bioinformatics (https://www.bioinformatics.com.cn/) and R version 4.3.2 with the ggplot2 package to visualize the results.
“Herb–Compound–Target” Network Construction
We used Cytoscape 3.10.133 to create a network analysis diagram called the “DH-HQ-compound-potential target” that graphically represents the connection between the medicine and the illness. The graphic depicts the probable active chemicals and effect targets of DH-HQ, with nodes representing these compounds and targets and the edges representing the interactions between them.
MR Analysis
The Genome-Wide Association Study (GWAS) pooled dataset encompasses a vast repository of information on the relationships between diseases and genetic variants. The integration of Mendelian randomization (MR) into contemporary computational biology has strengthened causal inference by utilizing genetic variants as instrumental variables to minimize bias from confounders. This approach capitalizes on the random segregation of alleles during meiosis, which inherently reduces susceptibility to environmental, lifestyle, or socioeconomic influences that typically obscure observational associations. Crucially, the temporal precedence of genetic polymorphisms—existing prior to disease manifestation—provides robust evidence for causal directionality distinct from reverse causation.34
We leveraged this dataset to further explore the impact of the intersecting genes previously mentioned on ARDS and conducted a two-sample MR analysis to identify specific genes within the gene profile that exert a causal effect on ARDS. The exposure data were derived from the gene expression quantitative trait loci (eQTL) dataset in the IEU Open GWAS project (https://gwas.mrcieu.ac.uk/; version 8.3.0), which comprises data from 31,684 individuals and 19,942 transcripts.35 We excluded SNPs with a P-value>0.5, clumping distance <10,000 kb, linkage disequilibrium (LD) r² >0.001, and F-statistics<10, and used the remaining SNPs as the exposure files for further analyses.36,37 Detailed data can be found in Supplementary Tables 1 and 2. The outcome data for ARDS were sourced from the Finnish database (https://finngen.gitbook.io/documentation), with GWAS ID “finngen_R10_J10_ARDS”, encompassing 357 ARDS cases and 406,536 controls (https://storage.googleapis.com/finngen-public-data-r10/summary_stats/finngen_R10_J10_ARDS.gz). All data are derived from European ancestry populations and are publicly available, thus requiring no additional ethical approval or informed consent (Supplementary Tables 1 and 2).
We performed the MR causal analysis on the combined exposure and outcome data using the “TwoSampleMR” R package,38 while also assessing heterogeneity, pleiotropy, and sensitivity. Five commonly used methods—Inverse Variance Weighted (IVW), MR Egger, Weighted Median, Simple Mode, and Weighted Mode—were employed for this MR analysis, with IVW and Weighted Median serving as the primary techniques for establishing a causal relationship between genes and disease. Heterogeneity was evaluated using Cochran’s Q test, with a P-value less than 0.05 indicating heterogeneity. Pleiotropy, which examines whether the instrumental variables influence the outcome solely through exposure, was assessed via the MR-Egger intercept and MR-PRESSO methods, with P-values of <0.05 indicating pleiotropy.39 A sensitivity analysis was conducted to evaluate the robustness of MR estimates, including a “leave-one-out” analysis, where each instrumental variable is excluded one by one to observe its influence on the overall causal effect of individual SNPs.40 Finally, scatter plots, funnel plots, and forest plots were generated to visually represent the MR analysis results.
Correlation Between Hub Genes and Immune Cells
CIBERSORT, a deconvolution algorithm often referred to as “digital cytometry”, enables the precise quantification of 22 distinct immune cell types based on gene expression datasets and performs immune cell infiltration analyses through linear support vector regression.41,42 In this study, we employed the CIBERSORT algorithm via the R package to analyze immune cell infiltration in the gene expression profiles of patients with ARDS, aiming to determine the relative composition of immune cells in their peripheral blood. Given the sample size of 49 (N<50), we utilized the Shapiro–Wilk test to assess the normality of the data distribution and employed Spearman’s rank correlation coefficient to evaluate the non-parametric correlations between target genes and immune cell types, considering a P-value of <0.05 statistically significant. To visually present the findings, we used the ggplot2, ggcorrplot, and ggpubr packages to generate visualizations, including lollipop plots, box plots, and heatmaps, providing a clear and intuitive display of the analysis results.
Single-Cell Analysis
We conducted an analysis of two human PBMC single-cell datasets, GSE151263 and GSE167363.43,44 The GSE151263 dataset comprises sequencing data from four patients with sepsis and three patients diagnosed with “sepsis + ARDS”, whereas the GSE167363 dataset contains samples from two healthy controls, two non-surviving sepsis patients, and three sepsis survivors. For the purpose of comparison, we specifically selected the “sepsis + ARDS” group from GSE151263 and the healthy control group from GSE167363.
During the preprocessing stage, rigorous data integration and quality control procedures were employed, whereby we retained only high-quality cells exhibiting feature counts between 200 and 2,500 and a mitochondrial gene percentage below 5%. Subsequently, the gene expression data for each cell were normalized using the NormalizeData function, followed by the application of FindVariableFeatures, Harmony, PCA, and UMAP to rectify batch effects and ensure data consistency. Cell type annotation was performed based on well-established PBMC markers, and the initial annotations were further refined through reference mapping.45 To maintain analytical consistency and optimize downstream processing using the Seurat (v4.3.0) package, the cell count was standardized to 5,000 cells per dataset. Finally, the expression levels of previously identified hub genes were meticulously validated within the single-cell datasets, ensuring the robustness of our findings.
ROC and GSEA Analysis
To validate the diagnostic capacity of hub genes for disease severity and their prognostic value for survival outcomes, we utilized two independent external validation cohorts (GSE69063 and GSE95233). Given that pulmonary involvement is a primary complication of sepsis—often serving as the initial target of systemic inflammatory responses—early identification of patients progressing to ARDS is critical for prognosis. Diagnostic performance was quantified using ROC curves and area under the curve (AUC) values generated by the pROC package. Subsequently, multivariate logistic regression was employed to calculate odds ratios (ORs), 95% confidence intervals, and p-values for associations between hub gene expression and survival status, with results visualized via forest plots to delineate their prognostic utility.
Using the original discovery cohort (GSE243066), Pearson correlation analysis was performed to assess pairwise correlations (coefficient R) between hub genes and their candidate targets, with statistical significance (p) determined. Interactions were visualized through scatter plots integrated with linear regression models. Genes showing significant correlations with hub genes were subjected to GSEA via the clusterProfiler package to identify biologically relevant pathways, with stringent criteria (p<0.05) applied for functional annotation. All R codes are provided in Supplementary File 1.
Molecular Docking Between Emodin-Baicalein and Hub Genes
Molecular docking is a computational simulation method that studies molecular interactions. By molecular docking, the binding modes of molecules can be predicted, which provides guidance for drug design. This method enables the estimation of the binding site and affinity of a ligand and a protein.46,47 To further elucidate the relationship and mechanism of action between the active ingredients of DH-HQ and its core targets, we conducted molecular docking studies to assess the interaction strength between the receptors and ligands. Based on the most active compounds found in Rhizoma Rhei and Scutellaria baicalensis Georgi, baicalein and emodin were selected as ligands, while hub genes were chosen as the target proteins for molecular docking simulations.48,49 For the receptor protein crystal structure derived from X-ray diffraction, we used the PDB database50 (https://www.rcsb.org) to obtain the required information. For the ligand, we used TCMSP to get the information in the mol2 format, or OpenBabelGUI51 to convert the sdf format from PubChem to the mol2 format. Next, we used AutoDockTools 1.5.6, AutoDock4,52 AutoGrid4, and PyMOL2.2 to visualize and analyze the docking outcomes. Throughout the docking procedure, we executed preliminary actions including water elimination, hydrogenation, charge calculation, and the establishment of Grid Boxes for receptors and ligands.
Experiment Validation
Animal Experiments and Model Establishment
We purchased 45 C57BL/6 mice aged 6–8 weeks and weighing 23±3 g from Shanghai JieSiJie Laboratory Animal Co. Ltd. The mice were housed under controlled conditions with a temperature of 26 ± 3 °C, relative humidity of 60–70%, and a 12-hour light/dark cycle, with access to food and water ad libitum. After a one-week acclimation period, the mice were randomly divided into the following five groups of nine mice: the NC group, LPS group, LPS+DH (emodin) group, LPS+HQ (baicalein) group, and LPS+DH-HQ (emodin+baicalein) group.
ALI Model Establishment: The mice were intraperitoneally injected with 0.5% sodium pentobarbital (10 µg/g) for anesthesia. Once fully anesthetized, mice in the LPS, LPS+DH, LPS+HQ, and LPS+DH-HQ groups were administered LPS (concentration: 1.25 µg/µL) intranasally, with 20 µL of the LPS solution gently applied to the nasal tip in two consecutive doses to avoid excessive aspiration and coughing, totaling 40 µL. Mice in the NC group were administered an equal volume of phosphate-buffered saline (PBS).
Emodin (Lot. No.: S30748) and baicalein (Lot. No.: B20571) were procured from Shanghai Yuanye Bio-Technology Co., Ltd. Based on the classical formulations for ALI and sepsis treatment, “Liang Ge San” and “San Huang Xie Xin Tang”, the emodin-to-baicalein ratio was determined to be 2:1.53,54 Three grams of emodin and baicalein were pre-dissolved in 600 mL of 0.5% sodium carboxymethyl cellulose and dimethyl sulfoxide (DMSO), preparing monomer solutions at a concentration of 1 mg/200 µL. A mixture of 200 mL of the emodin solution and 100 mL of the baicalein solution was used to formulate the DH-HQ suspension. Mice in the LPS+DH, LPS+HQ, and LPS+DH-HQ groups were administered the respective drug suspensions (40 mg/kg) via oral gavage in two doses within 2–12 hours post-ALI induction, with an interval of ≥8 hours between doses.55–57Humane endpoints were pre-defined to minimize suffering, including: severe respiratory distress (labored breathing or cyanosis), prolonged immobility (>24 hours), or inability to access food/water. Animals meeting any endpoint criterion were immediately euthanized via intraperitoneal injection of sodium pentobarbital overdose (150 mg/kg). All procedures were designed to reduce pain and distress.
Body Weight and Clinical Score
Throughout the study, the body weight and clinical scores of the mice were meticulously monitored on a daily basis, before and after model induction. Clinical scoring was based on the following five distinct criteria: weight loss exceeding 10%, the appearance of rash, ocular discharge, labored respiration, and lethargy, with each parameter assigned a score of 1, culminating in a maximum possible score of 5. At the point where the most pronounced differences in body weight and clinical scores were observed, left lung tissues were harvested for hematoxylin and eosin (H&E) staining.
Lung Histopathology
After harvesting lung tissues from the mice, they were thoroughly rinsed with PBS, and any excess moisture was blotted dry with filter paper. The tissues were then fixed in 4% paraformaldehyde (PFA). The following day, the samples were sent to Wuhan Servicebio Technology Co., Ltd. for histopathological processing, including fixation, embedding, and paraffin sectioning. The paraffin sections were first deparaffinized in an eco-friendly dewaxing solution and absolute ethanol, followed by rehydration. Frozen sections were warmed and fixed, after which high-definition consistent staining pretreatment was performed. The sections were then stained with hematoxylin, followed by differentiation and bluing. Subsequently, the sections were dehydrated in 95% ethanol and stained with eosin. Finally, they were cleared through sequential dehydration in absolute ethanol and n-butanol and mounted using neutral balsam. Upon completion of these procedures, the sections were examined under a microscope for image acquisition and analysis.
Statistical Analysis
Quantitative data are presented as the mean ± standard error of the mean. Sample size justification: The animal group size (n=9 per group) was determined based on previous studies of LPS-induced ALI models in mice and our pilot experiments showing detectable treatment effects within this range.58 While formal power analysis was not conducted due to the exploratory nature of this therapeutic combination, our sample size aligns with recommendations for preclinical lung injury studies in rodents.59 The primary effect sizes for key outcomes (eg, reduction in inflammatory markers and histopathological scores) observed in pilot experiments suggested adequate detection power given the established variance in LPS models. The two-way ANOVA was employed for multiple-group comparisons with Tukey’s post-hoc test.
In the systems pharmacology analysis, KEGG pathway enrichment was assessed via the hypergeometric test; differentially expressed genes were analyzed using Welch’s t-test (unpaired two-sample, non-equal variance); Mendelian randomization (MR) prioritized the Inverse Variance Weighted (IVW) and Weighted Median methods to infer causal gene-disease relationships, supplemented by Cochran’s Q-test for heterogeneity and MR-Egger/PRESSO for horizontal pleiotropy evaluation; immune infiltration correlations were determined through Shapiro–Wilk normality testing and Spearman’s rank correlation coefficients; clinical diagnostic performance was quantified via ROC curve-derived AUC values, while multivariate logistic regression (odds ratios [ORs], 95% CIs) and forest plots were employed to delineate prognostic associations. All statistical analyses were conducted using R software and GraphPad Prism. Statistical significance was established for P-values below 0.05 (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Ethical Statement
All experimental protocols involving animals were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee (IACUC) of the Tenth Affiliated Hospital of Southern Medical University (Dongguan People’s Hospital) under approval number IACUC-AWEC-202403113.
This research utilizes publicly available, fully de-identified data, thus exempt from institutional ethics re-approval under Article 32 (Clause 1) of the Measures for Ethical Review of Life Science and Medical Research Involving Human Subjects (National Health Commission of China, effective February 18, 2023). Importantly, this study did not involve interference with public social behaviors, nor any activities related to human germ cells, embryos, reproductive cloning, chimeric research, or heritable genetic modifications, as explicitly prohibited under Chapter III (Risk Classification) of the aforementioned regulation.
Results
Active Ingredients and Potential Targets of DH-HQ
By applying the filtering criteria (DL and OB), 207 active constituents of the medications were extracted from the TCMSP, BATMAN-TCM, and ETCM databases. Among these, 116 were obtained from DH, 89 from HQ, and two from both databases. In addition, we acquired 301, 937, and 145 possible targets of DH, and 443, 486, and 227 potential targets of HQ, respectively, from these three datasets. Upon removing the duplicated components and targets shared by DH and HQ, we successfully identified 1551 possible targets of the herb pair. These targets are visually shown in Figure 2A.
Collection of Gene Targets in ALI
Through our searches in the NCBI-gene, OMIM, GeneCards, and DisGeNET databases, we identified 325, 994, 1,499, and 102 genes, respectively, associated with “ALI and ARDS”. Specifically, we identified 1,419 potential targets for ALI and 1,576 for ARDS. After combining and deduplicating these results, we presented a total of 2,462 potential disease gene targets (Figure 2B). In the GSE243066 dataset, a total of 4,239 differentially expressed genes were identified.
Therapeutic Targets of DH-HQ for ALI
Following a Venn diagram analysis to identify potential gene targets common to the medicine and the disease, we identified 93 overlapping genes (Figure 2C). These shared genes were considered pivotal targets, likely mediating the therapeutic effects of DH-HQ in the treatment of ALI. In conjunction with subsequent experimental results, we conducted a heatmap analysis of the 93 intersecting genes (Figure 2D), followed by transcriptomic validation of the selected hub genes LTB4R, MMP9, and POLR1B. The transcriptional expression patterns were consistent with the findings from the single-cell analyses (Figure 2E), further corroborating their significance (Supplementary Tables 3, 4 and 8–11).
PPI Network of DH-HQ Against ALI
We imported the shared targets of DH-HQ, ALI, and the GSE243006 dataset into the STRING database and generated the corresponding PPI network of overlapping targets. In this network, the edges represent interactions between nodes, with a higher number of edges indicating stronger interactions. The PPI network comprised 93 nodes and 819 edges, with an average node degree of 17.6. The data file, downloaded from STRING in the TSV format, was further analyzed using Cytoscape 3.10.1, where the targets were ranked based on their degree values. Ultimately, a comprehensive set of 93 key genes implicated in the therapeutic effect of DH-HQ on ALI was identified, with the top five genes—GAPDH, IL1B, IL10, MMP9, and TP53—emerging as core targets based on their degree ranking (Figure 3; Supplementary Tables 5 and 6).
GO and KEGG Pathway Enrichment Analyses of DH-HQ Against ALI
Using the DAVID database, we conducted GO and KEGG pathway enrichment analyses for 93 key targets. We found a total of 2038 GO terms, which consisted of 1668 biological processes (81.8%), 107 cellular components (5.3%), 263 molecular functions (12.9%), and 211 KEGG pathways (P<0.05, FDR<0.05). Figures 4 demonstrates that the DH-HQ drug primarily interacts with protease, a chemokine receptor, a transition metal ion, a cytokine receptor, and C-C chemokine binding. These interactions are involved in the positive regulation of cytokine production, cellular response to lipids, positive regulation of intracellular signal transduction, regulation of cell population proliferation, and the cellular response to an oxygen-containing compound. The cellular components primarily consisted of a secretory granule lumen, sarcolemma, caveola, plasma membrane raft, and platelet alpha granule, among others.
The treatment of ALI using the DH-HQ drug involves several signal pathways, as illustrated in Figure 5. These pathways are involved in various biological processes, such as cancer, atherosclerosis, and virus infection. They include transcriptional misregulation in cancer, viral protein interaction with cytokine and cytokine receptor, the p53 signaling pathway, cytokine-cytokine receptor interaction, FoxO signaling pathway, HIF-1 signaling pathway, and NF-kappa B signaling pathway. Figure 5 displays the bijective relationship between pathways and genes as well as their relevance (Supplementary Tables 7 and 12).
Drug-Compounds-Targets Network
Following the examination of the active components of the medications, we established a network including the identified substances and targets. As seen in Figure 6: the “Drug-Compounds-Targets” network, which has not undergone screening, consists of 227 nodes and 828 edges. These nodes represent medications, drug components, and their related targets. The color of a node is determined by its degree value, with darker colors indicating higher degree values. Every edge symbolizes the connection between the “drug-drug active ingredient” and the “active ingredient-target”. The left side of the diagram depicts DH and its constituent elements as pink circles, while the right side represents HQ and its constituent elements as pink circles. In the center, there are two tiny pink nodes, A1 and A2, which symbolize the shared compounds of DH and HQ. The remaining green circles indicate the target genes.
MR Result
Leveraging the eQTL database with selection criteria of P <0.5, clumping distance < 10,000 kb, LD r² > 0.001, and F-statistic > 10, we performed the MR analysis on the 93 previously identified genes to explore their potential causal associations with ARDS. Following rigorous heterogeneity, pleiotropy, and sensitivity analyses, three genes were identified as having a negative causal relationship with ARDS. Using the IVW method for the most robust causal inference, elevated levels of LTB4R, Matrix metalloproteinase-9 (MMP9), and RNA polymerase I subunit RPA2 (POLR1B) were found to significantly decrease the risk of ARDS. Specifically, the OR [95% CI] for LTB4R was 0.615 [0.405–0.933] (P = 0.022), for MMP9 it was 0.657 [0.441–0.978] (P = 0.038), and for POLR1B it was 0.712 [0.519–0.977] (P = 0.035) (Figure 7). These robust causal associations suggest that elevated expression of LTB4R, MMP9, and POLR1B may exert protective effects against ARDS pathogenesis, potentially through modulating leukotriene-mediated inflammatory responses (LTB4R), extracellular matrix remodeling in alveolar injury (MMP9), and ribosomal biogenesis-linked cellular stress adaptation (POLR1B), thereby establishing their roles as plausible therapeutic targets supported by genetic evidence.
Immune Infiltration Relevance of 3 Hub Genes
We utilized the CIBERSORT algorithm to conduct immune infiltration analyses on the GSE243066 dataset. Remarkably, patients with ARDS exhibited elevated levels of neutrophils and M0 macrophages compared with healthy individuals. The MR analysis further revealed that MMP9 and LTB4R were positively associated with M0 macrophages and neutrophils. Specifically, MMP9 correlated with M0 macrophages at 0.73 and with neutrophils at 0.61, while LTB4R showed correlations of 0.43 with M0 macrophages and 0.84 with neutrophils. In contrast, POLR1B exhibited negative correlations, with coefficients of −0.35 for M0 macrophages and −0.52 for neutrophils (Figure 8). This coordinated pro-inflammatory pattern (MMP9/LTB4R promoting myeloid cell infiltration) and anti-inflammatory counterbalance (POLR1B suppressing neutrophil/macrophage accumulation) may reflect a dynamic immunoregulatory axis in ARDS, where dysregulated metalloproteinase activity and leukotriene signaling drive tissue-destructive inflammation, while ribosomal biosynthesis genes like POLR1B potentially mitigate cellular stress-induced immune hyperactivation.
Single-Cell Sequencing Results
Leveraging cell surface markers and reference mapping results, we annotated the clusters generated at a resolution of 0.5 using the FindClusters function, identifying 17 distinct cell populations (Supplementary Figure 1). During the analysis of cell communication, we identified that the TNFSF14-LTBR and LTA-(LTB+LTBR) receptor-ligand interactions play significant roles in the progress of ARDS. The TNFSF14-LTBR interaction exerts a conductive effect among various cell types, primarily regulating the interaction between CD16 monocytes and plasmacytoid dendritic cells (pDCs) with conventional dendritic cells (cDCs), thereby enhancing antiviral responses and activating immune cells. Meanwhile, the LTA-(LTB+LTBR) signaling pathway, involving lymphotoxin (LT), is widely present in the autocrine and paracrine signaling between T cells and dendritic cells (DCs). This pathway is crucial for the formation and maintenance of lymphoid structures, as it can induce apoptosis and inflammatory responses, playing an essential role in the activation of the immune system (Figure 9).
To validate hub gene expression in peripheral blood monocytes, we examined the differential expression of hub genes between ARDS patients and healthy controls. Notably, LTB4R expression was significantly elevated, whereas MMP9 and POLR1B expression levels were decreased. In ARDS patients, the average expression of LTB4R in peripheral blood macrophages was 0.13, compared to 0.09 in the healthy group (P = 0.0163 < 0.05). MMP9 was exclusively expressed in ARDS (P = 8.24e−09 < 0.0005). Likewise, POLR1B expression was unique to the ARDS group, with an expression level of 0.02 (P = 0.00437 < 0.005) (Figure 10). The correlation with MR analysis findings suggests: This phenotypic paradox may arise from disease stage-specific regulatory mechanisms—the acute upregulation of LTB4R in ARDS monocytes reflects tissue injury-driven compensatory anti-inflammatory feedback (adapting to dynamic disease progression), while MR-revealed genetically inherent LTB4R overexpression provides sustained protection against systemic hyperinflammation through enhanced innate immune homeostasis (at the lifelong risk level). This highlights the essential divergence between context-induced expression (single-cell evidence) and constitutionally determined expression (genetic evidence) in pathogenic regulation.
Diagnostic and Prognostic Potential of Hub Genes in ARDS and LTB4R-Related GSEA Pathways
We performed ROC analysis and prognostic modeling for sepsis-associated ALI, revealing distinct predictive capacities of hub genes across datasets. In the GSE69063 cohort, LTB4R (AUC=0.716), MMP9 (AUC=0.711), and POLR1B (AUC=0.73) demonstrated robust diagnostic performance for sepsis severity, with high specificity and sensitivity, suggesting their clinical utility in guiding ALI diagnosis. In contrast, within the GSE95233 cohort, only POLR1B exhibited significant predictive power (AUC=0.75), supported by prognostic modeling via nomogram analysis showing a statistically significant association with survival outcomes (OR=0.04, p<0.05) (Figure 11).
Further analysis revealed inverse correlations between POLR1B and LTB4R/MMP9 (p<0.05), while LTB4R and MMP9 showed strong positive co-expression (p<0.05) (Figure 12A). To elucidate the biological role of LTB4R—a key mediator of cellular communication—we conducted GSEA analysis on its correlated gene targets. Top enriched pathways included IL-6/JAK/STAT3 signaling, interferon responses, mTORC1-mediated metabolic reprogramming, and TNF-α-driven inflammation (via NF-κB-dependent barrier disruption and Fas/FasL-mediated alveolar epithelial apoptosis),60 collectively establishing LTB4R as a hub for immunometabolic dysregulation in ARDS pathogenesis (Figure 12B).
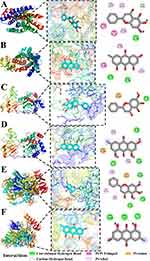
Molecular Docking Verification
The receptor-ligand docking hypothesis states that the binding energy determines the affinity between the ligand and the receptor, and the binding strength is negatively correlated with docking energy. The lower the binding energy, the greater the likelihood of contact between the ligand and receptor. The binding energy values between all receptors and ligands are less than −7 kcal/mol, with the lowest affinity binding energy being −7.3 kcal/mol while the highest affinity binding energy reaching −8.8 kcal/mol. This suggests that these key compounds are crucial in the treatment of ALI (Figure 13).
We utilized molecular docking techniques to generate visual representations of the interactions between the compounds and the key genes. In these pairings, the binding energy between LTB4R and baicalein is calculated at −8.6 kcal/mol. Figure 13A illustrates that this ligand forms Pi-Pi T-shaped interactions with PHE-275 and TRP-234, while Pi-Alkyl bonds are established at VAL-67, CYS-97, and ALA-274. Additionally, a Carbon–Hydrogen bond is observed at HIS-94, and a Conventional Hydrogen bond at GLU-185. Conversely, the binding energy between LTB4R and emodin measures −7.3 kcal/mol, as depicted in Figure 13B. Emodin forms Pi-Alkyl interactions with PHE-169, PRO-170, and ILE-271, alongside Conventional Hydrogen bonds at ARG-178 and ARG-156. When baicalein binds to MMP9, the interaction presents a binding energy of −7.5 kcal/mol, as shown in Figure 13C, characterized by Pi-Anion bonds at ASP-182 and a Conventional Hydrogen bond at ASN-38. The emodin-MMP9 interaction, which exhibits a binding energy of −8.7 kcal/mol (Figure 13D), involves Pi-Anion bonds at ASP-182, Pi-Alkyl interactions at ARG-51 and LEU-44, and Conventional Hydrogen bonds at ASN-38, ASP-185, and TYR-52.The interaction between baicalein and POLR1B manifests the highest binding affinity, with an energy of −8.8 kcal/mol. This robust binding is mediated through the formation of Conventional Hydrogen bonds at THR-906, ARG-85, and TYR-113, as illustrated in Figure 13E. In comparison, the binding energy of emodin with POLR1B is slightly lower, at −8.1 kcal/mol. Emodin engages in Pi-Alkyl interactions with LYS-1704, LEU-12, and ARG-11, while a Pi-Anion bond is observed at ASP-107. Furthermore, the complex is stabilized by the formation of Conventional Hydrogen bonds at ASN-1060, ASP-108, and GLN-109, as depicted in Figure 13F. (Supplementary Table 13)
Experimental Verification
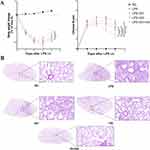
Post-model induction, the body weight variations and clinical scores of the mice were recorded daily at a fixed time. During the first three days, the mice exhibited progressive weakness, characterized by continuous weight loss and an increase in clinical scores. However, a marked turning point was observed on day four, when all mice began to show signs of recovery, reflected by a gradual increase in body weight and a reduction in clinical scores. This improvement was most pronounced in the DH-HQ+LPS group. On day four, the intergroup difference in body weight between the LPS and DH-HQ+LPS groups was statistically significant, with a P-value of 0.0328 (P < 0.05). Furthermore, the clinical score differences on day four revealed highly significant intergroup variations, with P-values of <0.0001 for LPS + DH-HQ vs LPS, 0.0005 for LPS + DH-HQ vs LPS + DH, and 0.0028 for LPS + DH-HQ vs LPS + HQ (P < 0.005) (Figure 14A).
To evaluate the effects of emodin and baicalein on the lung tissue structure of ALI mice, we employed H&E staining to analyze the morphological and histological changes. This assessment included the integrity and morphological alterations of alveoli, such as alveolar wall thickening, the presence of hemorrhage, or exudate in the alveolar spaces, the quantity and distribution of inflammatory cells like neutrophils and macrophages, interstitial edema, as well as signs of hemorrhage and fibrosis. As illustrated in Figure 14B, intranasal PBS administration did not induce significant structural changes in the lung tissue. However, LPS-induced ALI exhibited pronounced inflammatory cell infiltration, substantial thickening of the alveolar walls, and increased pulmonary blood flow. Treatment with emodin or baicalein alone reversed these pathological changes. Notably, the combined administration of emodin and baicalein considerably ameliorated the severity of lung lesions in ALI mice. The full spectrum of pathological findings is provided in Supplementary Figure 2. While these findings demonstrate therapeutic potential, we acknowledge that the cohort size (n=9 per group) imposes limitations on statistical generalizability, necessitating validation in larger-scale preclinical studies to confirm translational relevance.
Discussion
With a fatality rate of up to 38.5%, ALI is a clinical emergency marked by bilateral diffuse edema and refractory hypoxemia.3 Serious acute pancreatitis, viral pneumonia, and extrapulmonary sepsis are among the conditions that might cause it.61,62 Medicinal treatments for ALI are primarily classified as either physiological or anti-inflammatory based on the pathophysiological cause of the illness. Various drugs that intervene in different stages of inflammation are the mainstays of anti-inflammatory therapy. These drugs include protease inhibitors, antiproteases, ibuprofen, corticosteroids, pharmaconutrients, complement inhibitors, matrix metalloprotease modification, ketoconazole, and anti-oxidants. Physiology-based pharmacological therapies primarily focus on interventions for the patient’s ventilation, perfusion, and diffusion processes. More specifically, colony-stimulating factors, stem cells, and growth factors all play a role.63–67 Researchers have been attempting to discover a new medication to treat ALI for a long time; however, none of them has shown any promise in clinical trials that could provide patients with long-term relief due to the severity of the disease.68 The development of drugs based on natural compounds has consistently served as a major catalyst for novel drug research. Recently, researchers have examined various effects of natural substances in the treatment of ALI, identifying compounds that possess lung-protective properties and exhibit several anti-inflammatory effects. These compounds include alkaloids, flavonoids, baicalein, wogonin, emodin, and terpenoids.69 Our study focused on the traditional Chinese herbs DH and HQ, through which we identified and screened 207 potent and reliable bioactive compounds. At the microscopic level, the binding targets of hundreds of bioactive compounds exhibit extensive complexity. Through comprehensive searches of online databases, we identified 1,551 predicted binding targets. However, not all of these targets are directly relevant to the pathogenesis of ALI. Consequently, we conducted a rigorous screening process, narrowing the focus to those specifically involved in the disease process of ALI, ultimately identifying 93 intersecting genes. The PPI network analysis revealed that GAPDH, IL1B, IL10, MMP9, and TP53 occupy central positions within the interaction network, playing pivotal regulatory roles.
TCM has a long-standing tradition spanning thousands of years in the effective treatment of pulmonary diseases. Chinese medicine, a crucial component of TCM, serves as a valuable resource of natural products. It plays an irreplaceable and essential function in the adjuvant treatment of ALI. DH and HQ are an often-utilized herb pair in TCM to address ALI.1,11 According to TCM philosophy, DH is believed to have pharmacological actions that involve clearing heat and purging fire, promoting bowel movement and blood circulation.70 Laboratory studies have established that DH contains several natural components, including aloe emodin, rhein, emodin, and chrysophanol, which have demonstrated anti-inflammatory, antibacterial, and anti-fibrotic properties. Additionally, these components have the ability to prevent ALI progression.71 For instance, according to the empirical findings of Yang Q et al,56 emodin had a substantial impact on the unique miRNA expression profile of exosomes, consequently enhancing ALI resulting from severe acute pancreatitis. A study conducted by Fei Jiang et al72 verified that the DH extract known as chrysophanol protects mice from ALI caused by Klebsiella pneumoniae. The effects of this protective effect include lowering the level of reactive oxygen species in lung macrophages, decreasing immune cell infiltration, and improving the survival rate of the mice. This finding underscores the effectiveness of chrysophanol in mitigating ALI by inhibiting pro-inflammatory factors. HQ is a root-derived medicinal substance with a complex composition. It has various constituents that act on multiple targets and produce multiple effects. The flavonoid extract, which is a crucial element, has been scientifically demonstrated to possess anti-inflammatory, anti-complement, and anti-viral activities when used to treat ALI.73 The study conducted by Yu Long et al74 showcased the efficacy of the baicalin liposome in reducing the lung damage score, decreasing the W/D ratio, and suppressing pro-inflammatory variables. The potential mechanism by which it improves lipopolysaccharide (LPS)-induced ALI involves the activation of the JNK-ERK and TLR4-NFκBp65 signaling pathways. In this study, we collected the therapeutic targets for ALI/ARDS and DH-HQ from established disease databases, subsequently constructing a “drug-component-target” network consisting of 132 compounds and 93 core targets. This network illustrates that the pharmacological effects of these compounds are mediated through a sophisticated interplay of multi-target and multi-pathway interactions, underscoring the complex mechanisms of action of these herbal constituents. In addition to well-characterized compounds such as wogonin, baicalein, and emodin, several less-explored molecules—including norwogonin, progesterone, epiberberine, and 5,7,2,6-tetrahydroxyflavone—present intriguing potential for anti-ALI activity, though their mechanisms remain largely undefined. Future investigations should focus on these understudied compounds to explore novel avenues for ALI intervention and therapy, with the goal of identifying innovative agents that exhibit therapeutic efficacy against ALI.
Leukotrienes, pro-inflammatory lipid mediators derived from arachidonic acid through the action of 5-lipoxygenase (5-LO), play a critical role in neutrophil chemotaxis. Leukotriene B4 (LTB4) is generated by the hydrolytic cleavage of leukotriene A4 (LTA4) via LTA4 hydrolase.75 LTB4 exerts its effects by signaling through the receptors BLT1 and BLT2, inducing neutrophil adhesion to endothelial cells and thereby promoting chemotaxis. Previous studies have demonstrated elevated LTB4 levels in inflammatory bowel disease and ARDS.76 TNFSF14 mediates signaling through LTBR, thereby regulating both innate and adaptive immune responses. This signaling pathway plays a crucial role in the activation of immune cells, including the proliferation and survival of T cells and B cells. The LTBR signaling pathway is essential for the differentiation of various immune cell types and the maintenance of lymphoid organs, particularly in supporting the balance of dendritic cells and influencing the development of peripheral lymphoid structures.77–79Activation of the LT signaling pathway associated with LTBR enhances the proliferation and differentiation of T cells and B cells, promotes the development of lymphoid tissues, and regulates local inflammatory responses by influencing vascular permeability and the infiltration of inflammatory cells.80 A study by John et al on human pulmonary microvascular endothelial cells revealed that LTB4 and its metabolites can activate neutrophil oxidase, triggering their inflammatory response.81 MMP9, a key target in ALI research, is highly expressed in tumor invasion and various inflammatory conditions.82,83 As an inflammatory mediator, matrix metalloproteinases (MMPs) contribute to ALI by degrading alveolar endothelial and epithelial proteins, including basement membrane components, anchoring proteins, and intercellular junction proteins.84 In an immune complex model, MMP9-deficient mice exhibited reduced lung injury compared with their wild-type counterparts.85 Yao et al demonstrated that LPS stimulation upregulated MMP9 expression in mice, and the administration of recombinant MMP9 exacerbated LPS-induced macrophage polarization toward the M1 phenotype, a process that was suppressed by MMP9 siRNA intervention.86 POLR1B, the human homolog of the Rpa1 subunit within RNA polymerase I (Pol I), is essential for the synthesis of key ribosomal components and rRNA transcription.87 Although research on POLR1B in inflammation and ALI is limited, its role in lung cancer, particularly non-small cell lung cancer, has been implicated in the regulation of tumor cell proliferation, autophagosome assembly, and glucose uptake.88
Our PPI network incorporates extensively studied targets, such as TP53, IL-1B, IL-10, and MMP9, alongside less-explored targets like LTB4R and POLR1B, which have received limited attention in the treatment of ALI. MR analyses identified LTB4R, POLR1B, and MMP9 as being closely associated with ALI pathogenesis, not only exhibiting differential expression following pathological changes in ALI but also contributing as causative factors in its development. Immune infiltration and single-cell analyses in ARDS further confirmed the differential expression of these hub genes within macrophages, suggesting their crucial roles in immune regulation.
The identified hub genes (LTB4R, MMP9, POLR1B) demonstrated significant clinical value in the diagnosis and prognosis of sepsis-associated ALI, with POLR1B exhibiting particularly robust stratification capability. Crucially, these genes exhibited significant mutual regulatory relationships, suggesting a coordinated mechanism underlying their therapeutic efficacy. GSEA analysis revealed that LTB4R primarily modulates immunoregulatory pathways such as cytokine-cytokine receptor interaction and chemokine signaling, highlighting its central role in reprogramming immune-inflammatory crosstalk during ALI progression. This multi-gene synergy not only resolves inflammatory imbalance but also orchestrates systemic immune homeostasis, thereby positioning these hub genes as strategic therapeutic targets for precision intervention in ALI-associated immune dysregulation. Consequently, we propose that by modulating both these well-established and understudied targets, the DH-HQ herbal pair may considerably influence the pathological progression of ALI and promote the repair of lung tissues in the disease.
Through the utilization of GO/KEGG pathway enrichment analyses on 93 key gene targets, we have determined that the DH-HQ herb pair antagonizes ALI primarily by affecting shared signal transduction pathways, including the lipid and atherosclerosis signaling pathway, transcriptional misregulation in cancer, the p53 signaling pathway, cytokine-cytokine receptor interaction, the NF-kappa B signaling pathway, the HIF-1 signaling pathway, and the FoxO signaling pathway. Nevertheless, the involvement of pathways including the lipid and atherosclerosis and fluid shear stress signaling pathway is uncommon in the pathogenesis of ALI. Several molecular functions and biological processes, including the positive regulation of cytokine production, the positive regulation of intracellular signal transduction, and the inflammatory response, may be accomplished by regulating the key complex nodes in these pathways via the application of DH-HQ herb pairs. We posit that DH-HQ exerts its regulatory effects on ALI pathogenesis through a multitude of complex pathways and intricate interactions, underscoring its multifaceted therapeutic potential in modulating this disease.
To validate the predictions from network pharmacology and MR analyses, we employed molecular docking simulations to examine the interactions between key hub genes and the commonly studied compounds, emodin and baicalein. The docking results revealed that these compounds exhibited strong binding affinities toward disease-related targets. Notably, the binding energy between POLR1B and baicalein was the highest among all interactions, with the binding energies of other compound–target pairs consistently below −7.3 kcal/mol, indicating considerable ligand–receptor activity. Furthermore, beyond the three targets and two compounds illustrated in the Figures, other natural medicinal constituents may also demonstrate substantial binding affinities with these targets. The potential effects of these additional compounds and targets should not be overlooked. Our foundational experiments revealed that DH-HQ exhibits considerable therapeutic potential in the treatment of ALI. Compared with the traditional single-compound therapies using emodin or baicalein, the combination of these two compounds demonstrated a more pronounced and synergistic effect. This aligns with the TCM principle of compound synergy, where multi-target therapies involving the combination of various compounds may offer enhanced efficacy in disease amelioration. Such an approach suggests that DH-HQ’s multi-component strategy holds greater promise for effective ALI treatment than individual compounds alone.
Therefore, we propose the following hypothesis: The DH-HQ herb pair protects against ALI through a tripartite network targeting LTB4R, MMP9, and POLR1B. Key components include: (1) DH-HQ active compounds (eg, emodin, baicalein) block LTB4R-mediated neutrophil chemotaxis, suppressing leukocyte-endothelial adhesion and cytokine storm initiation; (2) MMP9 inhibition mitigates alveolar basement membrane degradation and antagonizes M1 macrophage polarization, synergistically controlling acute inflammation and structural damage; (3) The novel MR-identified target POLR1B likely modulates rRNA synthesis to reprogram immune cell metabolism and cooperates with TP53 signaling to sustain alveolar epithelial survival/regeneration. Our study breaks the mold of traditional ALI network pharmacology by shifting focus from classical anti-inflammatory pathways (NF-κB/TNF-α) and broad synergy to an inflammation-repair equilibrium model. We pioneer POLR1B—a ribosomal biogenesis driver of alveolar repair (validated via single-cell/Mendelian randomization)—defying apoptosis/oxidative stress-centered frameworks, and reposition LTB4R as a cytokine storm trigger (supported by GSEA/docking) rather than its previously uncharacterized auxiliary signaling role. These insights collectively reconstruct ALI pathogenesis through an integrated inflammation-repair equilibrium model.
Conclusion
This study aimed to elucidate and predict the action mechanism of the DH-HQ herb pair, commonly utilized in TCM for ALI treatment. Leveraging network pharmacology platforms rooted in bioinformatics and big data analytics, the investigation revealed that the DH-HQ herb pair addresses ALI through the modulation of various molecular entities, including Serotonin, Stigmasterol, and baicalein. The study further identified critical interactions with key proteins, including POLR1B, LTB4R, and MMP9, which play pivotal roles in inflammation and immune response. These interactions contribute to gene expression regulation and the activation of protein kinases, ultimately leading to the observed therapeutic effects.
This study highlights the challenge of accurately assessing the active components in the herbal compound owing to its complex composition. Database analyses may lead to erroneous findings, and the presence of common flavonoids in the compound might interact with other tests, leading to false-positive outcomes. While our MR analysis strengthened causal inferences between targets and ALI outcomes, key methodological constraints—including unresolved vertical pleiotropy and insufficient correction for population stratification—may introduce bias in causal effect estimation. Furthermore, given the extensive range of compounds and gene targets involved, our molecular docking analysis was focused solely on the interactions between three key hub genes and the compounds (emodin and baicalein). This selective approach, while insightful, represents only a fraction of the potential interactions within the broader therapeutic landscape. As a result, we were unable to investigate all the other crucial targets and compounds. These limitations necessitate additional clarification regarding the effectiveness of DH-HQ couples in this treatment process through in vitro and in vivo investigations, as well as pharmacokinetic approaches.
Future research should focus on studying the natural substances found in traditional Chinese medicine, as TCM might have notable benefits in the treatment of ALI. However, the absence of high-level clinical randomized controlled trials could restrict TCM use. Our current findings, derived entirely from preclinical computational models and molecular simulations, must undergo phased toxicity profiling and efficacy validation through standardized animal models before initiating human trials. Furthermore, given that certain components possess carcinogenic and mutagenic toxicity, it is imperative to exercise caution when employing them in clinical settings.
Abbreviations
ALI, Acute lung injury; ARDS, acute respiratory distress syndrome; PICU, pediatric intensive care unit; MMP9, Matrix metalloproteinase-9; LTB4, Leukotriene B4; POLR1B, RNA polymerase I subunit RPA2; TCM, Traditional Chinese Medicine; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein-protein interaction; ROC, receiver operating characteristic; GSEA, Gene Set Enrichment Analysis; ADMET, absorption, distribution, metabolism, excretion and toxicity; DL, drug-likeness; OB, oral bioavailability; FDA, Food and Drug Administration; OMIM, Online Mendelian Inheritance in Man; tsv, tab-separated values; EC, eigenvector; CC, closeness centrality; BC, betweenness centrality; BP, biological process; CC, cell component; MF, molecular function; GWAS, Genome-Wide Association Study; MR, Mendelian randomization; eQTL, expression quantitative trait loci; IVW, Inverse Variance Weighted; LD, linkage disequilibrium; KP, Klebsiella pneumoniae; LPS, Lipopolysaccharide; PBS, phosphate-buffered saline; H&E, hematoxylin and eosin; PFA, paraformaldehyde; RCTs, Randomized Controlled Trials; NSCLC, non-small cell lung cancer; 5-LO, 5-lipoxygenase.
Data Sharing Statement
The datasets generated during and analysed during the current study are available in the TCMSP database (https://old.tcmsp-e.com/tcmsp.php), BATMAN-TCM database (http://bionet.ncpsb.org.cn/batman-tcm/index.php), ETCM database (http://www.tcmip.cn/ETCM/index.php/Home/Index/), NCBI-gene database (https://www.ncbi.nlm.nih.gov/gene/), GEO database (http://www.ncbi.nlm.nih.gov/geo/), STRING database (https://string-db.org/), IEU Open GWAS project (https://gwas.mrcieu.ac.uk/) and Finnish database (https://finngen.gitbook.io/documentation). Clinical trial number: not applicable.
Acknowledgments
We sincerely appreciate the TCMSP, ETCM, BATMAN, Swiss Institute of Bioinformatics, GeneCards, OMIM, DisGeNET, UniProt, PubChem, STRING, Venny 2.1, jvenn, Cytoscape, DAVID database, PDB database OpenBabelGUI, CB-Dock2, AutoDockTools 1.5.6, AutoDock4, AutoGrid4, PyMOL2.2 and R software for data collection, statistical analysis, and figure plotting. We thank Shanghai NewCore Biotechnology Co., Ltd. (https://www.bioinformatics.com.cn, last accessed on 10 Nov 2023) for providing data analysis and visualization support.
Authorship Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital & Shenzhen Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Shenzhen (No.SZ2020ZD012); Supported by Sanming Project of Medicine in Shenzhen (No. SZSM202311003); Sponsored by National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital & Shenzhen Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Shenzhen (No. E010224009); Sponsored by National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital & Shenzhen Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Shenzhen (No. E0102240010); Sponsored by National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital & Shenzhen Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Shenzhen (No. E010224009). Supported by Shenzhen High-level Hospital Construction Fund; Supported by National Natural Science Foundation of China (No. 81971882); Supported by Suzhou Medical and Health Application Basic Research Project (SKJY2021079); Supported by Shanghai Engineering Research Center of Peri-operative Organ Support and Function Preservation (20DZ2254200). Supported by the Fundamental Research Funds for the Central Universities (No. 24×010202059).
Disclosure
The authors declare that there are no conflicts of interest in this work.
References
1. An Y, Zhang H, Wang R, et al. Biomarkers, signaling pathways, and programmed cell death in acute lung injury and its treatment with traditional Chinese medicine: a narrative review. Eur Rev Med Pharmacol Sci. 2023;27(21):10157–10170. doi:10.26355/eurrev_202311_34292
2. Khemani RG, Smith L, Lopez-Fernandez YM, et al. Paediatric acute respiratory distress syndrome incidence and epidemiology (PARDIE): an international, observational study. Lancet Respir Med. 2019;7(2):115–128. doi:10.1016/S2213-2600(18)30344-8
3. Rubenfeld GD, Caldwell E, Peabody E, et al. Incidence and outcomes of acute lung injury. N Engl J Med. 2005;353(16):1685–1693. doi:10.1056/NEJMoa050333
4. Qian G, Fang H, Chen A, et al. A hub gene signature as a therapeutic target and biomarker for sepsis and geriatric sepsis-induced ARDS concomitant with COVID-19 infection. Front Immunol. 2023;14:1257834. doi:10.3389/fimmu.2023.1257834
5. Sun Z, Chen A, Fang H, et al. B cell-derived IL-10 promotes the resolution of lipopolysaccharide-induced acute lung injury. Cell Death Dis. 2023;14(7):418. doi:10.1038/s41419-023-05954-2
6. Zheng L, Chen S, Cao Y, et al. Combination of comprehensive two-dimensional prostate cancer cell membrane chromatographic system and network pharmacology for characterizing membrane binding active components from Radix et Rhizoma Rhei and their targets. J Chromatogr A. 2018;1564:145–154. doi:10.1016/j.chroma.2018.06.015
7. Li J, Li T, Li Z, Song Z, Gong X. Nephroprotective mechanisms of Rhizoma Chuanxiong and Radix et Rhizoma Rhei against acute renal injury and renal fibrosis based on network pharmacology and experimental validation. Front Pharmacol. 2023;14:1154743. doi:10.3389/fphar.2023.1154743
8. Xiang L, Gao Y, Chen S, Sun J, Wu J, Meng X. Therapeutic potential of Scutellaria baicalensis Georgi in lung cancer therapy. Phytomedicine. 2022;95:153727. doi:10.1016/j.phymed.2021.153727
9. Zhou X, Fu L, Wang P, Yang L, Zhu X, Li CG. Drug-herb interactions between Scutellaria baicalensis and pharmaceutical drugs: insights from experimental studies, mechanistic actions to clinical applications. Biomed Pharmacother. 2021;138:111445. doi:10.1016/j.biopha.2021.111445
10. Jang JY, Im E, Kim ND. Therapeutic potential of bioactive components from Scutellaria baicalensis Georgi in inflammatory bowel disease and colorectal cancer: a review. Int J Mol Sci. 2023;24(3):1954. doi:10.3390/ijms24031954
11. Li YJ, Jia YP, Zhang TT, et al. Study on the medication rules of traditional Chinese medicine in the treatment of acute lung injury and acute respiratory distress syndrome based on literature. World Chinese Med. 2020;15(12):1756–1761.
12. Ge P, Luo Y, Zhang J, et al. Mechanism investigation and clinical retrospective evaluation of Qingyi granules: pancreas cleaner about ameliorating severe acute pancreatitis with acute respiratory distress syndrome. Drug Des Devel Ther. 2024;Volume 18(18):2043–2061. doi:10.2147/DDDT.S454180
13. Li W, Jiang H, Zhang W, et al. Mechanisms of action of Sappan lignum for prostate cancer treatment: network pharmacology, molecular docking and experimental validation[J]. Front Pharmacol. 2024;15(15):1407525. doi:10.3389/fphar.2024.1407525
14. Jain B, Raj U, Varadwaj PK. Drug target interplay: a network-based analysis of human diseases and the drug targets. Curr Top Med Chem. 2018;18(13):1053–1061. doi:10.2174/1568026618666180719160922
15. Zhou Z, Chen B, Chen S, et al. Applications of network pharmacology in traditional Chinese medicine research. Evid Based Compl Alternat Med. 2020;2020(1):1646905. doi:10.1155/2020/1646905
16. Fu S, Zhou Y, Hu C, Xu Z, Hou J. Network pharmacology and molecular docking technology-based predictive study of the active ingredients and potential targets of rhubarb for the treatment of diabetic nephropathy. BMC Complement Med Ther. 2022;22(1):210. doi:10.1186/s12906-022-03662-6
17. Pagadala NS, Syed K, Tuszynski J. Software for molecular docking: a review. Biophys Rev. 2017;9(2):91–102. doi:10.1007/s12551-016-0247-1
18. Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6(1):13. doi:10.1186/1758-2946-6-13
19. Kong X, Liu C, Zhang Z, et al. BATMAN-TCM 2.0: an enhanced integrative database for known and predicted interactions between traditional Chinese medicine ingredients and target proteins. Nucleic Acids Res. 2023;52(D1):D1110–20.
20. Xu HY, Zhang YQ, Liu ZM, et al. ETCM: an encyclopaedia of traditional Chinese medicine. Nucleic Acids Res. 2019;47(D1):D976–D982. doi:10.1093/nar/gky987
21. Kim S. Getting the most out of PubChem for virtual screening. Expert Opin Drug Discov. 2016;11(9):843–855. doi:10.1080/17460441.2016.1216967
22. Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47(W1):W357–W364. doi:10.1093/nar/gkz382
23. Bateman A, Martin M-J, Orchard S, UniProt Consortium. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49(D1):D480–D489. doi:10.1093/nar/gkaa1100
24. Zhong Y, Hu L, Chen W, et al. Exploring the comorbidity mechanisms between asthma and idiopathic pulmonary fibrosis and the pharmacological mechanisms of Bu-Shen-Yi-Qi decoction therapy via network pharmacology[J]. BMC Compl Med Ther. 2022;22(1):151. doi:10.1186/s12906-022-03637-7
25. Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM.org: online Mendelian inheritance in man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43(Database issue):D789–98. doi:10.1093/nar/gku1205
26. Stelzer G, Rosen N, Plaschkes I, et al. The genecards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016;54(1):1.30.1–1.30.33. doi:10.1002/cpbi.5
27. Piñero J, Saüch J, Sanz F, Furlong LI. The DisGeNET cytoscape app: exploring and visualizing disease genomics data. Comput Struct Biotechnol J. 2021;19:2960–2967. doi:10.1016/j.csbj.2021.05.015
28. Mitsuyama Y, Matsumoto H, Togami Y, et al. T cell dysfunction in elderly ARDS patients based on miRNA and mRNA integration analysis. Front Immunol. 2024;15(15):1368446. doi:10.3389/fimmu.2024.1368446
29. Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. jvenn: an interactive Venn diagram viewer. BMC Bioinf. 2014;15(1):293. doi:10.1186/1471-2105-15-293
30. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi:10.1093/nar/gkw937
31. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi:10.1038/nprot.2008.211
32. Sherman BT, Hao M, Qiu J, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022;50(W1):W216–W221. doi:10.1093/nar/gkac194
33. Otasek D, Morris JH, Bouças J, Pico AR, Demchak B. Cytoscape automation: empowering workflow-based network analysis. Genome Biol. 2019;20(1):185. doi:10.1186/s13059-019-1758-4
34. Cao S, Gu Y, Lu G, et al. Causal correlations between plasma metabolites, inflammatory proteins, and chronic obstructive pulmonary disease: a Mendelian randomization and bioinformatics-based investigation. J Inflamm Res. 2025;Volume 18(18):4057–4073. doi:10.2147/JIR.S513526
35. Võsa U, Claringbould A, Westra HJ, et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat Genet. 2021;53(9):1300–1310. doi:10.1038/s41588-021-00913-z
36. Burgess S, Thompson SG. CRP CHD genetics collaboration. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. 2011;40(3):755–764. doi:10.1093/ije/dyr036
37. Burgess S, Dudbridge F, Thompson SG. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat Med. 2016;35(11):1880–1906. doi:10.1002/sim.6835
38. Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome[J]. Elife. 2018. doi:10.7554/eLife.34408
39. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–525. doi:10.1093/ije/dyv080
40. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32(5):377–389. doi:10.1007/s10654-017-0255-x
41. Chen B, Khodadoust MS, Liu CL, et al. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. 2018;1711(1711):243–259. doi:10.1007/978-1-4939-7493-1_12
42. Newman AM, Steen CB, Liu CL, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37(7):773–782. doi:10.1038/s41587-019-0114-2
43. Jiang Y, Rosborough BR, Chen J, et al. Single cell RNA sequencing identifies an early monocyte gene signature in acute respiratory distress syndrome. JCI Insight. 2020;5(13). doi:10.1172/jci.insight.135678
44. Qiu X, Li J, Bonenfant J, et al. Dynamic changes in human single-cell transcriptional signatures during fatal sepsis. J Leukoc Biol. 2021;110(6):1253–1268. doi:10.1002/JLB.5MA0721-825R
45. Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587.e29. doi:10.1016/j.cell.2021.04.048
46. Chen G, Seukep AJ, Guo M. Recent advances in molecular docking for the research and discovery of potential marine drugs. Mar Drugs. 2020;18(11):545. doi:10.3390/md18110545
47. Zhang X, Yang Y, Wen M, et al. Supplementary hesperidin alleviated CPT-11-induced diarrhea by modulating gut microbiota and inhibiting the IL-17 signaling pathway. J Agric Food Chem. 2025;73(10):5915–5930. doi:10.1021/acs.jafc.4c09602
48. Chen H, Zhang Y, Zhang W, et al. Inhibition of myeloid differentiation factor 2 by baicalein protects against acute lung injury. Phytomedicine. 2019;63(63):152997. doi:10.1016/j.phymed.2019.152997
49. Wu X, Yao J, Hu Q, et al. Emodin ameliorates acute pancreatitis-associated lung injury through inhibiting the alveolar macrophages pyroptosis. Front Pharmacol. 2022;13:873053. doi:10.3389/fphar.2022.873053
50. Burley SK, Bhikadiya C, Bi C, et al. RCSB protein data bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021;49(D1):D437–D451. doi:10.1093/nar/gkaa1038
51. O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open babel: an open chemical toolbox. J Cheminform. 2011;3(1):33. doi:10.1186/1758-2946-3-33
52. Seeliger D, de Groot BL. Ligand docking and binding site analysis with PyMOL and autodock/vina. J Comput Aided Mol Des. 2010;24(5):417–422. doi:10.1007/s10822-010-9352-6
53. Xiang L, Wang PM, Wang P, et al. Protective effects and mechanisms of rhei radix et rhizoma-scutellariae radix combination on hepatic inflammatory injury in endotoxemia model rats. Pharmacol Clini Chinese Mater Med. 2018;34(1):105–108.
54. Liu JX, Yu LZ, Wang L, et al. Protective effects of liangge powder on pulmonary edema in rats with acute lung injury induced by endotoxin. Pharmacol Clini Chinese Mater Med. 2012;28(3):2–5.
55. Xu C, Luo Y, Ntim M, et al. Effect of emodin on long non-coding RNA-mRNA networks in rats with severe acute pancreatitis-induced acute lung injury. J Cell Mol Med. 2021;25(4):1851–1866. doi:10.1111/jcmm.15525
56. Yang Q, Luo Y, Ge P, et al. Emodin ameliorates severe acute pancreatitis-associated acute lung injury in rats by modulating exosome-specific miRNA expression profiles. Int J Nanomed. 2023;18:6743–6761. doi:10.2147/IJN.S428924
57. Jiang C, Zhang J, Xie H, et al. Baicalein suppresses lipopolysaccharide-induced acute lung injury by regulating Drp1-dependent mitochondrial fission of macrophages. Biomed Pharmacother. 2022;145(145):112408. doi:10.1016/j.biopha.2021.112408
58. Zhu C, Chen J, Yan Z, et al. IL-22 alleviates sepsis-induced acute lung injury by inhibiting epithelial cell apoptosis associated with STAT3 signalling. J Inflamm Res. 2025;18(18):5383–5398. doi:10.2147/JIR.S496387
59. Jin S, Ding X, Yang C, et al. Mechanical ventilation exacerbates poly (I:C) induced acute lung injury: central role for caspase-11 and Gut-Lung axis. Front Immunol. 2021;12:693874. doi:10.3389/fimmu.2021.693874
60. Huang L, Du B, Cui X, et al. Nerelimomab alleviates capsaicin-induced acute lung injury by inhibiting TNF signaling and apoptosis. Pharmaceuticals. 2024;17(12):1694. doi:10.3390/ph17121694
61. Bos L, Ware LB. Acute respiratory distress syndrome: causes, pathophysiology, and phenotypes. Lancet. 2022;400(10358):1145–1156. doi:10.1016/S0140-6736(22)01485-4
62. Karki R, Kanneganti TD. The ‘cytokine storm’: molecular mechanisms and therapeutic prospects. Trends Immunol. 2021;42(8):681–705. doi:10.1016/j.it.2021.06.001
63. Impellizzeri D, Bruschetta G, Esposito E, Cuzzocrea S. Emerging drugs for acute lung injury. Expert Opin Emerg Drugs. 2015;20(1):75–89. doi:10.1517/14728214.2015.1000299
64. Sweeney RM, Griffiths M, McAuley D. Treatment of acute lung injury: current and emerging pharmacological therapies. Semin Respir Crit Care Med. 2013;34(4):487–498. doi:10.1055/s-0033-1351119
65. Yamashita CM, Lewis JF. Emerging therapies for treatment of acute lung injury and acute respiratory distress syndrome. Expert Opin Emerg Drugs. 2012;17(1):1–4. doi:10.1517/14728214.2012.667800
66. Zhang J, Ge P, Liu J, et al. Glucocorticoid treatment in acute respiratory distress syndrome: an overview on mechanistic insights and clinical benefit. Int J Mol Sci. 2023;24(15).
67. Qian G, Jiang W, Zou B, et al. LPS inactivation by a host lipase allows lung epithelial cell sensitization for allergic asthma. J Exp Med. 2018;215(9):2397–2412. doi:10.1084/jem.20172225
68. Fadanni GP, Calixto JB. Recent progress and prospects for anti-cytokine therapy in preclinical and clinical acute lung injury. Cytokine Growth Factor Rev. 2023;71-72:13–25. doi:10.1016/j.cytogfr.2023.07.002
69. He YQ, Zhou CC, Yu LY, et al. Natural product derived phytochemicals in managing acute lung injury by multiple mechanisms. Pharmacol Res. 2021;163:105224. doi:10.1016/j.phrs.2020.105224
70. Zhuang T, Gu X, Zhou N, Ding L, Yang L, Zhou M. Hepatoprotection and hepatotoxicity of Chinese herb Rhubarb (Dahuang): how to properly control the “General (Jiang Jun)” in Chinese medical herb. Biomed Pharmacother. 2020;127:110224. doi:10.1016/j.biopha.2020.110224
71. Cao YJ, Pu ZJ, Tang YP, et al. Advances in bio-active constituents, pharmacology and clinical applications of rhubarb. Chin Med. 2017;12(1):36. doi:10.1186/s13020-017-0158-5
72. Jiang F, Jiang J, He W, et al. Chrysophanol alleviates acute lung injury caused by Klebsiella pneumoniae infection by inhibiting pro-inflammatory cytokine production. Phytother Res. 2023;37(7):2965–2978. doi:10.1002/ptr.7792
73. Zhi HJ, Zhu HY, Zhang YY, Lu Y, Li H, Chen DF. In vivo effect of quantified flavonoids-enriched extract of Scutellaria baicalensis root on acute lung injury induced by influenza A virus. Phytomedicine. 2019;57:105–116. doi:10.1016/j.phymed.2018.12.009
74. Long Y, Xiang Y, Liu S, et al. Baicalin liposome alleviates lipopolysaccharide-induced acute lung injury in mice via inhibiting TLR4/JNK/ERK/NF-κB pathway. Mediators Inflamm. 2020;2020:8414062. doi:10.1155/2020/8414062
75. Murphy RC, Gijón MA. Biosynthesis and metabolism of leukotrienes[J]. Biochem J. 2007;405(3):379–395. doi:10.1042/BJ20070289
76. Sala A, Zarini S, Bolla M. Leukotrienes: lipid bioeffectors of inflammatory reactions. Biochemistry. 1998;63(1):84–92.
77. Han M, Sun Y, Zhao W, et al. Comprehensive characterization of TNFSF14/LIGHT with implications in prognosis and immunotherapy of human gliomas. Front Immunol. 2022;13(13):1025286. doi:10.3389/fimmu.2022.1025286
78. Riffelmacher T, Giles DA, Zahner S, et al. Metabolic activation and colitis pathogenesis is prevented by lymphotoxin β receptor expression in neutrophils. Mucosal Immunol. 2021;14(3):679–690. doi:10.1038/s41385-021-00378-7
79. Ware CF, Croft M, Neil GA. Realigning the LIGHT signaling network to control dysregulated inflammation. J Exp Med. 2022;219(7). doi:10.1084/jem.20220236
80. Fernandes MT, Ghezzo MN, Silveira AB, et al. Lymphotoxin-β receptor in microenvironmental cells promotes the development of T-cell acute lymphoblastic leukaemia with cortical/mature immunophenotype. Br J Haematol. 2015;171(5):736–751. doi:10.1111/bjh.13760
81. Eun JC, Moore EE, Banerjee A, et al. Leukotriene b4 and its metabolites prime the neutrophil oxidase and induce proinflammatory activation of human pulmonary microvascular endothelial cells. Shock. 2011;35(3):240–244. doi:10.1097/SHK.0b013e3181faceb3
82. Xu D, McKee CM, Cao Y, et al. Matrix metalloproteinase-9 regulates tumor cell invasion through cleavage of protease nexin-1. Cancer Res. 2010;70(17):6988–6998. doi:10.1158/0008-5472.CAN-10-0242
83. Zhang D, Zhang JT, Pan Y, et al. Syndecan-1 shedding by matrix metalloproteinase-9 signaling regulates alveolar epithelial tight junction in lipopolysaccharide-induced early acute lung injury. J Inflamm Res. 2021;14:5801–5816. doi:10.2147/JIR.S331020
84. Davey A, McAuley DF, O’Kane CM. Matrix metalloproteinases in acute lung injury: mediators of injury and drivers of repair. Eur Respir J. 2011;38(4):959–970. doi:10.1183/09031936.00032111
85. Warner RL, Lewis CS, Beltran L, et al. The role of metalloelastase in immune complex-induced acute lung injury. Am J Pathol. 2001;158(6):2139–2144. doi:10.1016/S0002-9440(10)64685-8
86. Tong Y, Yu Z, Chen Z, et al. The HIV protease inhibitor Saquinavir attenuates sepsis-induced acute lung injury and promotes M2 macrophage polarization via targeting matrix metalloproteinase-9. Cell Death Dis. 2021;12(1):67. doi:10.1038/s41419-020-03320-0
87. Andersen JS, Lam YW, Leung AK, et al. Nucleolar proteome dynamics. Nature. 2005;433(7021):77–83. doi:10.1038/nature03207
88. Yang F, Liu H, Zhao J, et al. POLR1B is upregulated and promotes cell proliferation in non-small cell lung cancer. Oncol Lett. 2020;19(1):671–680. doi:10.3892/ol.2019.11136
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.