Back to Journals » Journal of Inflammation Research » Volume 17
RNA Sequencing Reveals That the Genes Related to Cell Cycle and Glycolysis Play an Essential Role in IL-27-Induced Keratinocyte Hyperproliferation
Authors Wu Z, Wang R, Liu Y, Yang B , Wang H
Received 7 June 2024
Accepted for publication 26 October 2024
Published 4 November 2024 Volume 2024:17 Pages 8165—8180
DOI https://doi.org/10.2147/JIR.S481835
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tara Strutt
Zijun Wu,1,* Ruijing Wang,1,* Yuanjun Liu,1 Bin Yang,2 Huiping Wang1
1Department of Dermatovenereology, Tianjin Medical University General Hospital/Tianjin Institute of Sexually Transmitted Disease, Tianjin, People’s Republic of China; 2Department of Burn and Plastic Surgery, General Hospital of Central Theater Command, Wuhan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Bin Yang, Department of Burn and Plastic Surgery, General Hospital of Central Theater Command, Wuhan, Hubei, People’s Republic of China, Email [email protected] Huiping Wang, Department of Dermatovenereology, Tianjin Medical University General Hospital/Tianjin Institute of Sexually Transmitted Disease, Tianjin, People’s Republic of China, Email [email protected]
Background: Psoriasis is characterized by accelerated proliferation of epidermal keratinocytes. IL-27 is relevant to psoriasis pathogenesis. We previously found that IL-27 stimulates the proliferation of keratinocytes. However, the mRNAs involved in the process have not been fully studied. This study aims to identify potential pathways and hub genes associated with proliferation in keratinocytes with IL-27 intervention by bioinformatics analysis.
Methods: The mRNA expression profiles from HaCaT cells with or without IL-27 treated were analyzed by bioinformatics tools. The protein–protein interaction (PPI) network was constructed to screen gene clusters and hub genes associated with proliferation. Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Set Enrichment Analysis (GSEA) were used to identify the function of the mRNAs. The GEO database and quantitative real-time PCR (qPCR) were used to verify the expression levels of hub genes in psoriatic skin lesions and IL-27-treated psoriasiform keratinocytes, respectively.
Results: We found 1257 differentially expressed genes and screened 2 crucial gene clusters. GO analysis revealed that Cluster 1 was mainly enriched in “Mitotic sister chromatid segregation” and “Spindle”. Cluster 2 was mainly enriched in the “Pyruvate metabolic process” and “Oxidoreductase complex”. KEGG analysis showed that Cluster 1 and Cluster 2 were mainly enriched in “Cell cycle” and “Glycolysis/Gluconeogenesis”, respectively. We then identified 6 hub genes enriched in the two pathways, including CCNB1, PTTG1, CDC20, PLK1, PKM, and LDHA. GSEA complemented the role of the mitochondrial “Oxidative phosphorylation” pathway. Moreover, we found that 6 hub genes were upregulated in psoriasis skin lesions and IL-27 elevated the hub genes expression in M5-induced psoriasiform keratinocytes.
Conclusion: IL-27 possibly promotes glycolysis, mitochondrial oxidative phosphorylation, and cell cycle progression in keratinocytes. Additionally, we identified CCNB1, PTTG1, CDC20, PLK1, PKM, and LDHA as hub genes that may be involved in the mechanism of IL-27 facilitating keratinocyte proliferation in psoriasis.
Keywords: psoriasis, glycolysis, oxidative phosphorylation, bioinformatics
Introduction
Psoriasis is a common chronic relapsing papulosquamous dermatosis induced by the complicated interplay among immunity, genetics, and environment.1,2 The pathogenesis of psoriasis is associated with the complex crosstalk between immune cells and epidermal keratinocytes. There are many activated immune cells, especially T cells, dendritic cells, and macrophages, that infiltrate psoriatic skin lesions. Under the influence of internal and external factors, myeloid dendritic cells are activated and release cytokines such as IL-12 and IL-23. IL-12 induces Th1 cells to differentiate and release TNF-α and IFN-γ, while IL-23 induces Th17 and Th22 cells to differentiate and release IL-17, TNF-α and IL-22. These pro-inflammatory factors promote proliferation, abnormal differentiation, and secretion of multiple cytokines in keratinocytes, which activates and attracts more immune cells to the lesion, further exacerbating inflammation and leading to psoriasis.3,4 Among them, the accelerated proliferation and shortened mitotic cycle of keratinocytes in the basal layer of the epidermis are important pathophysiological features of psoriasis.5,6
As a member of the IL-12 cytokine family, IL-27 is a heterodimeric cytokine comprised p28 and Epstein-Barr virus-induced gene 3 subunits. IL-27 is primarily generated by activated antigen-presenting cells and activates downstream signaling by combining its receptor which consists of gp130 and IL-27Rα.7–9 IL-27 exerts both pro-inflammatory and anti-inflammatory effects on psoriasis. On the one hand, Shibata et al find that in psoriatic patients, serum IL-27 level is significantly increased and positively related to the severity of the disease.10 IL-27 increases IFN-γ levels in imiquimod-treated mouse and induces the production of CXCL9, CXCL10, and CXCL11 in keratinocytes. These chemokines selectively attract activated Th1 cells, leading to disease onset.9,10 On the other hand, Chen et al report that in patients with moderate to severe psoriasis, serum IL-27 level and skin lesional mRNA levels of IL-27p28, IL-27Rα, and gp130 are significantly decreased. IL-27 inhibits the secretion of IL-17 by CD4+ T cells and reduces the level of IL-17A in the mouse psoriasiform model.9 Additionally, IL-27 dose-dependently inhibits TNF-α-induced secretion of IL-1α and CCL20 in keratinocytes. IL-27 may act as an anti-inflammatory factor in the inflammation expansion phase of psoriasis.10 Our previous study found that IL-27 can drive keratinocyte proliferation.11 This may be one of the mechanisms by which IL-27 acts as a pro-inflammatory player in psoriasis. However, the mRNAs involved in the process have not been fully studied.
This study aims to identify potential pathways and hub genes associated with proliferation in keratinocytes with IL-27 intervention. The mRNA expression profiles from human immortalized keratinocytes (HaCaT cells) with or without IL-27 treated were analyzed. The protein–protein interaction (PPI) network was established to identify important gene clusters and hub genes associated with proliferation. Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Set Enrichment Analysis (GSEA) were performed to investigate the functions of mRNAs. Finally, the Gene Expression Omnibus (GEO) database and quantitative real-time PCR (qPCR) were used to verify the expression levels of hub genes in psoriatic skin lesions and IL-27-treated psoriasiform keratinocytes, respectively. This study will provide new insight into how IL-27 facilitates the proliferation of keratinocytes in psoriasis.
Materials and Methods
Cell Culture
HaCaT cells (CL-0090, Procell, China) were grown in MEM (G4550-500ML, Servicebio, China) with 15% FBS and 1% P/S and were incubated at 37°C with 5% CO2. Cells in the logarithmic growth stage were collected for subculture and further experimentation. In our previous study, HaCaT cell viability was significantly increased and approached maximum after being stimulated by 50 ng/mL IL-27 (200–38, PEPROTECH, USA) for 24 h. Therefore, 50 ng/mL and 24 h were selected as the optimal conditions for IL-27 intervention in RNA sequencing or subsequent experiments. The cells were divided into the IL-27 group (50 ng/mL IL-27) and the control group (0 ng/mL IL-27). M5 is a cytokine cocktail to construct a psoriasiform cell model at a concentration of 10 ng/mL for 24 hours. It consists of IL-17A (200–17), IL-22 (200–22), IL-1α (200–01A), TNF-α (300–01A), and oncostatin M (300–10), which we all purchased from PEPROTECH, USA.
RNA Extraction and qPCR
Following the manufacturer’s instructions, total RNAs of HaCaT cells were isolated using RNAiso Plus (9108, TaKaRa, China) and reverse transcribed into cDNA using the PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) (RR047A, TaKaRa, China). The cDNA was then amplified by PCR using TB Green® Premix Ex Taq™ II (Tli RNaseH Plus) (RR820A, TaKaRa, China) with the following primers: CCNB1 Forward: AATAAGGCGAAGATCAACATGGC, Reverse: TTTGTTACCAATGTCCCCAAGAG; CDC20 Forward: ATGCGCCAGAGGGTTATCAG, Reverse: GATACGGTCTGGCAGGGAAG; PTTG1 Forward: ACCCGTGTGGTTGCTAAGG, Reverse: ACGTGGTGTTGAAACTTGAGAT; PKM Forward: AAGGGTGTGAACCTTCCTGG, Reverse: GCTCGACCCCAAACTTCAGA; LDHA Forward: ATGGCAACTCTAAAGGATCAGC, Reverse: CCAACCCCAACAACTGTAATCT; PLK1 Forward: CCTGCACCGAAACCGAGTTAT, Reverse: CCGTCATATTCGACTTTGGTTGC; ACTB Forward: CATGTACGTTGCTATCCAGGC, Reverse: CTCCTTAATGTCACGCACGAT. The relative changes in mRNA expression levels were analyzed with the 2-ΔΔCt method and normalized to ACTB.
Differentially Expressed Genes Identification
The mRNA expression profiles of the IL-27 group and control group were from previously published data.11 The devtools package in the R software (version 4.2.3) was performed to draw a principal component analysis (PCA) plot. Log2|FC| ≥1.1 and q-value <0.05 were used as the threshold for screening differentially expressed genes (DE genes). The ggplot2 package in the R software was performed to draw a volcano plot to display the DE genes. Genes with -log10(q-value) ≥40 were labeled.
KEGG and GO Enrichment Analyses
KEGG and GO analyses were performed using the clusterProfiler package in the R software to explore the function of mRNAs. We regarded terms with p-value <0.05 as statistically significantly enriched. KEGG is used for signaling pathway analysis and GO is used for biological process (BP), cellular component (CC), and molecular function (MF) analyses. The KEGG pathway map was visualized by the KEGG Mapper – Color tool (https://www.kegg.jp/kegg/mapper/color.html).
PPI Network Construction
The STRING database (version 11.5) and Cytoscape software (version 3.8.0) were used to construct the PPI network. The minimum required interaction score is set to high confidence (0.700). The MCODE and cytoHubba plugins in Cytoscape software were performed to identify crucial gene clusters and hub genes, respectively. We defined clusters with scores greater than 6.5 and closely associated with cell proliferation as crucial gene clusters. The hub genes were identified by four algorithms (EPC, EcCentricity, Closeness, and Radiality) in the cytoHubba plug-in, which was visualized by the Draw Venn Diagram website (https://bioinformatics.psb.ugent.be/webtools/Venn/).
Gene Set Enrichment Analysis
The GSEA was performed to verify and supplement the results of the KEGG analysis. After preparing the expression dataset file (gct format) and phenotype labels file (cls format), we imported them into the GSEA software (version 4.3.3). The analysis parameters are set as follows. The gene sets database was selected as “c2.cp.kegg_medicus.v2023.2.Hs.symbols.gmt” and the number of permutations was 1000. The permutation type was gene_set and the metric for ranking genes was log2_Ratio_of_Classes. |NES| >1, nominal p-value <0.05 and FDR q-value <0.25 indicated statistically significant results.
GEO Database Analysis
Open access Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) database was used to analyze the levels of hub genes in psoriasis skin lesions. GSE14905 and GSE13355 datasets comprise normal skin from healthy controls and non-lesional skin and lesional skin from psoriasis patients. Raw data were processed using the transcripts per million (TPM) method and then transformed by normalization using log2 (TPM+1). Data analysis and visualization were performed using the ggplot2 package in the R software.
Statistical Analysis
Data analysis and plotting were performed using GraphPad Prism (version 8.3.0). The results were presented as mean ± SEM. An independent-sample t-test was applied to identify significant differences between two groups. Kruskal–Wallis test was used to compare multiple groups and Dunn’s multiple comparison test was used for multiple comparisons. p-value <0.05 indicated statistically significant results.
Results
Differentially Expressed Genes Analysis and Functional Enrichment Analysis
To determine the effect of mRNAs in IL-27-related keratinocyte proliferation, the mRNA expression profiles from the IL-27 group and control group were analyzed. The PCA plot showed that the IL-27 group and control group could be distinguished (Supplementary Figure 1). We found that 1257 genes were statistically differentially expressed (Figure 1A). These included 310 up-regulated genes and 947 down-regulated genes. To discover the function of DE genes, KEGG and GO enrichment analyses were performed. The results showed that the up-regulated DE genes were mainly enriched in “Steroid biosynthesis”, “Cell cycle”, “Glycolysis/Gluconeogenesis”, “Cholesterol metabolic process” and “Oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor” (Figure 1B and C). The down-regulated DE genes were mainly concentrated in the “MAPK signaling pathway”, “Calcium signaling pathway”, “Calcium channel complex” and “DNA-binding transcription repressor activity” (Figure 1D and E).
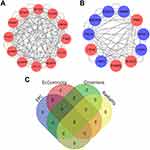
PPI Network Construction and Crucial Gene Clusters Screening
To identify important gene clusters in IL-27 facilitating the proliferation of keratinocytes, we established the PPI network for DE genes and then screened 2 crucial gene clusters related to cell proliferation. Cluster 1 (score = 11.455) included 12 nodes and 126 edges (Figure 2). GO analysis revealed that the BP term of Cluster 1 was involved in “Mitotic sister chromatid segregation” and “Mitotic nuclear division”, CC was involved in “Spindle” and “Kinetochore”, and MF was involved in “Microtubule binding” and “Tubulin binding” (Figure 3). Cluster 2 (score = 6.6) consisted of 11 nodes and 66 edges (Figure 2). Based on the GO analysis, the BP term of Cluster 2 was involved in the “Ribonucleotide metabolic process” and “Pyruvate metabolic process”, CC was involved in the “Oxidoreductase complex” and “Vesicle lumen”, and MF was involved in “Oxidoreductase activity, acting on CH-OH group of donors” and “Carbohydrate binding” (Figure 4). KEGG analysis showed that Cluster 1 and Cluster 2 were mainly associated with “Cell cycle” (Figure 3) and “Glycolysis/Gluconeogenesis” (Figure 4), respectively.
Hub Genes Analysis
To find out hub genes relevant to IL-27-stimulated proliferation of keratinocytes, we further explored the genes enriched in the “Cell cycle” and “Glycolysis/Gluconeogenesis” pathway. According to the results of the KEGG analysis, 4 genes enriched in the “Cell cycle” pathway of Cluster 1, containing CCNB1, PTTG1, PLK1, and CDC20 (Figure 3D). Additionally, 7 genes enriched in the “Glycolysis/Gluconeogenesis” pathway of Cluster 2, including PGAM4, HKDC1, LDHC, ALDOC, PKM, PKLR and LDHA (Figure 4D). According to the results of four algorithms (EPC, EcCentricity, Closeness, and Radiality) in the cytoHubba plug-in, the top 6 hub genes were selected from these 11 genes, consisting of CCNB1, PTTG1, CDC20, PLK1, PKM, and LDHA (Figure 2) (Table 1).
 |
Table 1 The Information of 6 hub Genes |
Gene Set Enrichment Analysis
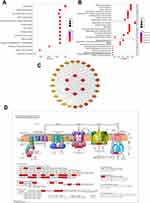
Next, to validate the results of the KEGG analysis, a GSEA was conducted. The results showed that the enriched pathways concentrated in “Mitochondrial complex UCP1 in thermogenesis”, “Electron transfer in complex I”, “Origin unwinding and elongation”, “Nuclear export of mRNA”, “DNA replication licensing” and “Glycolysis” (Figure 5). The GSEA not only verified the role of the “Glycolysis” and “Cell cycle” pathways but also revealed the role of mitochondria in IL-27 driving the proliferation of keratinocytes.
 |
Figure 5 The results of GSEA for mRNAs profiles from HaCaT cells with or without IL-27 treated. |
Bioinformatics Analysis of Mitochondria-Related Genes
To further investigate mitochondrial function in IL-27 facilitating the proliferation of keratinocyte, KEGG and GO analyses were performed for 31 up-regulated core genes enriched in “Mitochondrial complex UCP1 in thermogenesis” or “Electron transfer in complex I”, based on the results of GSEA. The results showed that the KEGG pathway mainly involved “Oxidative phosphorylation” (Figure 6A). GO BP involved specifically “Oxidative phosphorylation” and “Mitochondrial ATP synthesis coupled electron transport”. CC mainly contained “Mitochondrial inner membrane” and “Respiratory chain complex”. MF is mainly enriched in “Electron transfer activity”, “Oxidoreduction-driven active transmembrane transporter activity”, and “NADH dehydrogenase (ubiquinone) activity” (Figure 6B). Subsequently, the above 31 genes were used to construct a PPI network. These genes were ranked by the EPC algorithm. The top 6 genes were singled out (Figure 6C). Among these, NDUFAB1, NDUFA2, NDUFA10, NDUFA6, and NDUFS2 were enriched in NADH dehydrogenase. The 26 genes enriched in “Oxidative phosphorylation” were visualized in the KEGG pathway map (Figure 6D) (Table 2).
 |
Table 2 The Information on Genes Enriched in the Oxidative Phosphorylation Pathway |
Confirmation of Hub Genes Using qPCR
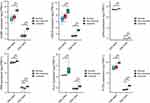
The transcriptional changes of hub genes were detected by qPCR. The results indicated that the expression levels of CCNB1 (P < 0.05), PTTG1 (P < 0.05), CDC20 (P < 0.001), PLK1 (P < 0.001), PKM (P < 0.001), and LDHA (P < 0.001) were significantly increased in the IL-27 group when compared with the control group, which was in line with the bioinformatics analysis (Figure 7).
 |
Figure 7 Verification of hub genes by qPCR. qPCR results of 6 hub genes: CCNB1, CDC20, LDHA, PKM, PLK1, and PTTG1. Data is displayed as mean ± SEM. *P < 0.05, ***P < 0.001. |
Expression Levels of Hub Genes in Psoriatic Skin Lesions
To analyze the expression of hub genes in psoriasis skin lesions, we used the GEO database. GSE14905 and GSE13355 comprise non-lesional skin and lesional skin from psoriasis patients and normal skin from healthy controls. The results showed that the levels of CCNB1, PTTG1, CDC20, PLK1, PKM, and LDHA in psoriasis lesional skin were significantly higher than in normal and non-lesional skin (P < 0.001) (Figure 8).
IL-27 Elevated Hub Genes Expression in M5-Induced Psoriasiform Keratinocytes
To explore the effects of IL-27 on hub genes in psoriatic keratinocytes, we induced psoriasiform inflammation in HaCaT cells with an M5 cocktail. The results indicated that IL-27 elevated the expression levels of CCNB1 (P < 0.001), PTTG1 (P < 0.05), CDC20 (P < 0.001), PLK1 (P < 0.001), PKM (P < 0.01), and LDHA (P < 0.001) in M5-induced keratinocyte (Figure 9).
Discussion
In this study, we obtained 1257 DE genes in IL-27-treated keratinocytes. By using PPI, GO, and KEGG analyses, we found that pathways of “Cell cycle” and “Glycolysis” were essential for IL-27 facilitating keratinocyte proliferation. Then, we identified 6 hub genes enriched in the two pathways, including CCNB1, PTTG1, CDC20, PLK1, PKM, and LDHA. The GSEA complemented the role of mitochondrial “Oxidative phosphorylation” in IL-27 facilitating the proliferation of keratinocytes.
Moreover, we found that 6 hub genes were upregulated in psoriasis skin lesions and IL-27 elevated the hub genes expression in M5-induced psoriasiform keratinocytes.
The balance between cell proliferation and differentiation is essential for maintaining epidermis homeostasis. Enhanced proliferation and attenuated differentiation are concomitant with the occurrence of hyperplastic skin disorders, such as psoriasis. This process depends on the interplay of multiple regulators involved in the cell cycle.12 In the present study, the cell cycle signaling pathway was discovered associated with IL-27-induced proliferation of keratinocytes. Meanwhile, we identified CCNB1, PTTG1, CDC20, and PLK1 as hub genes that were enriched in this pathway. CCNB1 (cyclin B1) is an indispensable regulator involved in the transition of G1/S and G2/M phase in the cell cycle. The enhanced level of Cyclin B1 appears to be characteristic of psoriasis.13 1,25-dihydroxyvitamin D3, which can inhibit CDC2 and cyclin B1 and thus lead to G2/M phase arrest in keratinocytes, is used as a therapeutic for psoriasis.14 PTTG1 is responsible for regulating the separation of sister chromatids and the transition from metaphase to anaphase.12 PTTG1 upregulation is associated with skin diseases characterized by increased proliferation and impaired differentiation in the epidermis.15 Studies have shown that PTTG1 is a susceptibility gene for psoriasis.16,17 In keratinocytes, there is a positive feedback loop between PTTG1 and TNF-α.15 Overexpressed PTTG1 also elevate cyclin B1. These cause anomalous proliferation and suppressed early differentiation in psoriasis skin lesions.12 CDC20 is involved in nucleus motion and chromosome separation at the metaphase–anaphase transition. Quek et al report that CDC20 merely exists in basal and suprabasal layers of the epidermis. Its mRNA abundance is 10-fold higher in human primary keratinocyte stem cells than in differentiated keratinocytes. Silencing CDC20 with siRNA reduces clonogenicity and stimulates differentiation.18 PLK1 is a serine/threonine kinase that has a vital role in regulating centrosome maturation, spindle assembly, chromosome segregation, and cytokinesis.19,20 Previous reports revealed that PLK1 increases in squamous cell carcinoma and basal cell carcinoma. In addition, the inhibition of PLK1 in the squamous cell carcinoma keratinocytes can lead to G2/M arrest, cell viability reduction, and apoptosis.21,22 Taken together, IL-27 possibly upregulates CCNB1, CDC20, PLK1, and PTTG1, facilitating cell cycle progression, to stimulate the proliferation of psoriatic keratinocytes.
Aerobic glycolysis is a sign of rapidly proliferating cells.23 Recently, an accelerated glycolytic rate has been revealed in psoriasis keratinocytes.24,25 According to our bioinformatics analysis, glycolysis was involved in IL-27 facilitating the proliferation of keratinocytes. In this pathway, PKM and LDHA were identified as hub genes. PKM is one of the rate-limiting enzymes in glycolysis, catalyzing the conversion of phosphoenolpyruvate to pyruvate. It consists of two isozymes: PKM1 and PKM2.26 Severely attenuated PKM1 expression by Ex9 mutation reducing keratinocytes proliferation.27 Likewise, lack of PKM2 inhibits keratinocyte proliferation and psoriasis inflammation in psoriatic mouse models.28 Additionally, in psoriasis epidermis, TRIM33 is found elevated and induces the ubiquitination and nuclear retention of PKM2, thus stimulating glycolysis and proliferation of keratinocytes.24 LDHA, one of the subunits of lactate dehydrogenase, significantly affects glycolytic metabolism by catalyzing the conversion of pyruvate to lactate. LDHA and lactate are highly expressed in psoriatic skin lesions.29–31 Luo et al report that keratin 17 and alpha-enolase are increased in the psoriasis epidermis. Their interplay drives keratin 17 translocating into the nucleus, which induces the transcription of LDHA and thus promotes the glycolysis and proliferation of keratinocytes.32 All these studies imply an essential role for PKM and LDHA in psoriasis pathogenesis. Therefore, it is likely that IL-27 induces the abnormal proliferation of psoriatic keratinocytes by regulating PKM and LDHA.
Based on the GSEA, KEGG, and GO analyses, in addition to enhancing glycolysis, IL-27 increased mitochondrial oxidative phosphorylation as well. Oxidative phosphorylation can supply energy and promote the synthesis of aspartate, which is conducive to cell proliferation.33 Increased mitochondrial oxidative phosphorylation level is found in the highly proliferative psoriatic keratinocytes.34–36 IL-17A, a well-known pathogenic factor of psoriasis, significantly enhances oxidative phosphorylation in keratinocytes.37,38 Studies have shown that ND1, ND2, ND4, ND5, and ND6, mitochondrial NADH dehydrogenase subunits that participate in the construction of complex I in the respiratory chain and contribute to electron transfer, are highly expressed in psoriasis skin lesions. The decoction of Jianpi-Yangxue-Jiiedu, a psoriatic effective therapeutic, diminishes their expression to decrease energy generated from oxidative phosphorylation, thus attenuating keratinocyte hyperproliferation.39 Similarly, in our findings, 5 mitochondria-associated genes with higher scores, including NDUFAB1, NDUFA2, NDUFA10, NDUFA6, and NDUFS2, were enriched in NADH dehydrogenase. These suggest the possibility that IL-27 drives the proliferation of keratinocytes by stimulating mitochondrial oxidative phosphorylation in the pathogenesis of psoriasis.
This study has several shortcomings. Firstly, whether the six hub genes we determined play a primary role in IL-27 facilitating the proliferation of keratinocytes remains to be explored. In addition, we did not observe the effects of IL-27 on proliferation-related pathways and genes in keratinocytes of the psoriasis animal model. Lastly, we did not perform functional experiments to further validate the identified pathways and genes.
Conclusion
IL-27 possibly promotes glycolysis, mitochondrial oxidative phosphorylation, and cell cycle progression in keratinocytes. In addition, we identified six hub genes, including CCNB1, PTTG1, CDC20, PLK1, PKM, and LDHA, highly expressed both in psoriatic lesions and IL-27-treated psoriasiform keratinocytes. Therefore, IL-27 may be involved in the pathogenesis of psoriasis by regulating these genes, but the specific molecular mechanism remains to be further studied.
Abbreviations
HaCaT cells, human-immortalized keratinocytes; PPI, protein–protein interaction; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; GSEA, Gene Set Enrichment Analysis; GEO, Gene Expression Omnibus; qPCR, quantitative real-time PCR; PCA, principal component analysis; DE genes, differentially expressed genes; BP, biological process; CC, cellular component; MF, molecular function; TPM, transcripts per million.
Data Sharing Statement
Any data in this study can be obtained from corresponding authors upon reasonable request.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was funded by Tianjin Key Medical Discipline (Specialty) Construction Project (No: TJYXZDXK-057B).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Zhou X, Chen Y, Cui L, Shi Y, Guo C. Advances in the pathogenesis of psoriasis: from keratinocyte perspective. Cell Death Dis. 2022;13(1):81. doi:10.1038/s41419-022-04523-3
2. Hawkes JE, Nguyen GH, Fujita M, et al. microRNAs in Psoriasis. J Investigative Dermatol. 2016;136(2):365–371. doi:10.1038/JID.2015.409
3. Armstrong AW, Read C. Pathophysiology, clinical presentation, and treatment of psoriasis: a review. JAMA. 2020;323(19):1945–1960. doi:10.1001/jama.2020.4006
4. Griffiths CEM, Armstrong AW, Gudjonsson JE, Barker J. Psoriasis. Lancet. 2021;397(10281):1301–1315. doi:10.1016/S0140-6736(20)32549-6
5. Grän F, Kerstan A, Serfling E, Goebeler M, Muhammad K. Current developments in the immunology of psoriasis. Yale J Biol Med. 2020;93(1):97–110. doi:10.1038/jid.2012.339
6. Thatikonda S, Pooladanda V, Sigalapalli DK, Godugu C. Piperlongumine regulates epigenetic modulation and alleviates psoriasis-like skin inflammation via inhibition of hyperproliferation and inflammation. Cell Death Dis. 2020;11(1):21. doi:10.1038/s41419-019-2212-y
7. Bosmann M, Ward PA. Modulation of inflammation by interleukin-27. J Leukocyte Biol. 2013;94(6):1159–1165. doi:10.1189/jlb.0213107
8. Villarino AV, Huang E, Hunter CA. Understanding the pro- and anti-inflammatory properties of IL-27. J Immunol. 2004;173(2):715–720. doi:10.4049/jimmunol.173.2.715
9. Chen W, Gong Y, Zhang X, et al. Decreased expression of IL-27 in moderate-to-severe psoriasis and its anti-inflammation role in imiquimod-induced psoriasis-like mouse model. J Dermatological Sci. 2017;85(2):115–123. doi:10.1016/j.jdermsci.2016.11.011
10. Shibata S, Tada Y, Kanda N, et al. Possible roles of IL-27 in the pathogenesis of psoriasis. J Investigative Dermatol. 2010;130(4):1034–1039. doi:10.1038/jid.2009.349
11. Wu Z, Yang Q, Xu K, Wu J, Yang B. Study on the key genes and molecular mechanisms of IL-27 promoting keratinocytes proliferation based on transcriptome sequencing. Clin Cosmet Invest Dermatol. 2023;16:1457–1472. doi:10.2147/CCID.S414633
12. Ishitsuka Y, Kawachi Y, Taguchi S, et al. Pituitary tumor-transforming gene 1 enhances proliferation and suppresses early differentiation of keratinocytes. J Investigative Dermatol. 2012;132(7):1775–1784. doi:10.1038/jid.2012.74
13. Miracco C, Pellegrino M, Flori ML, Vatti R, Materno M, Andreassi L. Cyclin D1, B and A expression and cell turnover in psoriatic skin lesions before and after cyclosporin treatment. British J Dermatol. 2000;143(5):950–956. doi:10.1046/j.1365-2133.2000.03826.x
14. Dai X, Yamasaki K, Yang L, et al. Keratinocyte G2/M growth arrest by 1,25-dihydroxyvitamin D3 is caused by Cdc2 phosphorylation through Wee1 and Myt1 regulation. J Investigative Dermatol. 2004;122(6):1356–1364. doi:10.1111/j.0022-202X.2004.22522.x
15. Ishitsuka Y, Kawachi Y, Maruyama H, et al. Pituitary tumor transforming gene 1 induces tumor necrosis factor-α production from keratinocytes: implication for involvement in the pathophysiology of psoriasis. J Investigative Dermatol. 2013;133(11):2566–2575. doi:10.1038/jid.2013.189
16. Sun LD, Cheng H, Wang ZX, et al. Association analyses identify six new psoriasis susceptibility loci in the Chinese population. Nature Genet. 2010;42(11):1005–1009. doi:10.1038/ng.690
17. Yang Q, Liu H, Qu L, et al. Investigation of 20 non-HLA (human leucocyte antigen) psoriasis susceptibility loci in Chinese patients with psoriatic arthritis and psoriasis vulgaris. British J Dermatol. 2013;168(5):1060–1065. doi:10.1111/bjd.12142
18. Quek LS, Grasset N, Jasmen JB, Robinson KS, Bellanger S. Dual role of the anaphase promoting complex/cyclosome in regulating stemness and differentiation in human primary keratinocytes. J Investigative Dermatol. 2018;138(8):1851–1861. doi:10.1016/j.jid.2018.02.033
19. Schmit TL, Ahmad N. Regulation of mitosis via mitotic kinases: new opportunities for cancer management. Mol Cancer Ther. 2007;6(7):1920–1931. doi:10.1158/1535-7163.MCT-06-0781
20. Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol. 2009;10(4):265–275. doi:10.1038/nrm2653
21. Schmit TL, Zhong W, Nihal M, Ahmad N. Polo-like kinase 1 (Plk1) in non-melanoma skin cancers. Cell Cycle. 2009;8(17):2697–2702. doi:10.4161/cc.8.17.9413
22. Watt SA, Pourreyron C, Purdie K, et al. Integrative mRNA profiling comparing cultured primary cells with clinical samples reveals PLK1 and C20orf20 as therapeutic targets in cutaneous squamous cell carcinoma. Oncogene. 2011;30(46):4666–4677. doi:10.1038/onc.2011.180
23. Abdel-Haleem AM, Lewis NE, Jamshidi N, Mineta K, Gao X, Gojobori T. The emerging facets of non-cancerous Warburg effect. Front Endocrinol. 2017;8:279. doi:10.3389/fendo.2017.00279
24. Yang L, Zhang J, Hu C, et al. Nuclear translocation of PKM2 mediates keratinocyte metabolic reprogramming in psoriasis. Experimental Dermatol. 2023;32(11):1960–1970. doi:10.1111/exd.14922
25. Kang H, Li X, Zhou Q, et al. Exploration of candidate biomarkers for human psoriasis based on gas chromatography-mass spectrometry serum metabolomics. British J Dermatol. 2017;176(3):713–722. doi:10.1111/bjd.15008
26. Demeter JB, Elshaarrawi A, Dowker-Key PD, Bettaieb A. The emerging role of PKM in keratinocyte homeostasis and pathophysiology. FEBS J. 2022;290(9):2311–2319. doi:10.1111/febs.16700
27. Nold SP, Sych K, Imre G, et al. Reciprocal abrogation of PKM isoforms: contradictory outcomes and differing impact of splicing signal on CRISPR/Cas9 mediates gene editing in keratinocytes. FEBS J. 2023;290(9):2338–2365. doi:10.1111/febs.16625
28. Liu YZ, Xu MY, Dai XY, et al. Pyruvate kinase M2 mediates glycolysis contributes to psoriasis by promoting keratinocyte proliferation. Front Pharmacol. 2021;12:765790. doi:10.3389/fphar.2021.765790
29. Zezina E, Sercan-Alp O, Herrmann M, Biesemann N. Glucose transporter 1 in rheumatoid arthritis and autoimmunity. Wiley Interdiscipl Rev Syst Biol Med. 2020;12(4):e1483. doi:10.1002/wsbm.1483
30. Wang Y, Qi C, Feng F, et al. Resveratrol ameliorates imiquimod-induced psoriasis-like mouse model via reducing macrophage infiltration and inhibiting glycolysis. J Inflamm Res. 2023;16:3823–3836. doi:10.2147/JIR.S416417
31. Lin HC, Chen YJ, Wei YH, et al. Lactic acid fermentation is required for NLRP3 inflammasome activation. Front Immunol. 2021;12:630380. doi:10.3389/fimmu.2021.630380
32. Luo Y, Pang B, Hao J, et al. Keratin 17 covalently binds to alpha-enolase and exacerbates proliferation of keratinocytes in psoriasis. Int J Bio Sci. 2023;19(11):3395–3411. doi:10.7150/ijbs.83141
33. Yao CH, Wang R, Wang Y, Kung CP, Weber JD, Patti GJ. Mitochondrial fusion supports increased oxidative phosphorylation during cell proliferation. eLife. 2019;8:e41351. doi:10.7554/eLife.41351
34. Liszewska A, Robak E, Bernacka M, Bogaczewicz J, Woźniacka A. Methotrexate use and NAD(+)/NADH metabolism in psoriatic keratinocytes. Postepy dermatologii i alergologii. 2020;37(1):19–22. doi:10.5114/ada.2020.93379
35. McGill A, Frank A, Emmett N, Turnbull DM, Birch-Machin MA, Reynolds NJ. The anti-psoriatic drug anthralin accumulates in keratinocyte mitochondria, dissipates mitochondrial membrane potential, and induces apoptosis through a pathway dependent on respiratory competent mitochondria. FASEB J. 2005;19(8):1012–1014. doi:10.1096/fj.04-2664fje
36. Zhu X, Zhang E, Qin L. The high expression of TOP2A and MELK induces the occurrence of psoriasis. Aging. 2024;16(4):3185–3199. doi:10.18632/aging.205519
37. Liu CT, Yen JJ, Brown DA, et al. Targeting Nrf2 with 3 H-1,2-dithiole-3-thione to moderate OXPHOS-driven oxidative stress attenuates IL-17A-induced psoriasis. Biomed Pharmacother. 2023;159:114294. doi:10.1016/j.biopha.2023.114294
38. Lee CL, Wang CM, Song YC, Liu CT, Chu MY, Yen HR. An alkaloid-rich phytopharmaceutical prepared from Qing Dai against IL-17A-induced psoriasis. J Ethnopharmacol. 2024;318(Pt A):116924. doi:10.1016/j.jep.2023.116924
39. Zhao N, Wang Y, Qu B, et al. Jianpi-Yangxue-Jiedu decoction improves the energy metabolism of psoriasis mice by regulating the electron transfer of oxidative phosphorylation. J Ethnopharmacol. 2024;324:117714. doi:10.1016/j.jep.2024.117714
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.