Back to Journals » OncoTargets and Therapy » Volume 18
Studying Immunogenic Cell Death in Human Colorectal Cancer Organoids
Authors Lisandrelli R, Winkler M , Trajanoski Z, Lamberti G
Received 20 February 2025
Accepted for publication 13 May 2025
Published 6 June 2025 Volume 2025:18 Pages 705—715
DOI https://doi.org/10.2147/OTT.S521346
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tohru Yamada
Rebecca Lisandrelli, Matthias Winkler, Zlatko Trajanoski, Giorgia Lamberti
Institute of Bioinformatics, Biocenter, Medical University of Innsbruck, Innsbruck, Austria
Correspondence: Giorgia Lamberti, Medical University of Innsbruck, Biocenter, Institute of Bioinformatics, Innrain 80, Innsbruck, 6020, Austria, Email [email protected]
Purpose: Combination therapies of chemotherapeutic agents and immunotherapy have shown promising results in the treatment of cold tumors. Various chemotherapies trigger immunogenic cell death (ICD) and release of hallmarks immunogenic damage associated molecular patterns (DAMPs) that have been related with immunostimulatory activities, leading to better patient prognosis. We aim to optimize in vitro assays to detect DAMPs release in response to chemotherapeutic agents using a colorectal cancer (CRC) organoids model.
Methods: CRC patient-derived organoids (PDOs) were treated either with oxaliplatin (OXA) or with 5-fluorouracil (5FU) and viability was measured with CellTiter-Glo3D Cell viability assay. Calreticulin (CALR) and high mobility group box 1 protein (HMGB1) intracellular translocation was assessed in immunofluorescence microscopy and quantified via co-localization with wheat germ agglutinin (WGA) and DAPI. Extracellular release of adenosine triphosphate (ATP) was quantified via specific luminescence assays.
Results: The results showed that CRC PDOs release DAMPs in a patient-specific manner in response to OXA and 5FU treatments.
Conclusion: This study successfully used immunofluorescence and luminescence methods to detect ICD-associated DAMPs release in CRC PDOs in response to chemotherapeutic treatments. This approach allows the recognition of patient-specific ICD activation and could help predict patient response to therapies.
Keywords: DAMPs, chemotherapy, combination therapies, OXA, 5FU, PDOs
Introduction
First described in 2005, immunogenic cell death (ICD) is a peculiar form of regulated cell death (RCD) able to activate an adaptive immune response in an immunocompetent environment.1,2 ICD can be triggered by a set of stimuli, such as pathogens and chemotherapeutic agents, resulting in the spatiotemporal emission of damage associated molecular patterns (DAMPs), giving rise to a stepwise process involving dendritic cell (DCs) recruitment, antigen processing, maturation, and antigen presentation to T cells.3,4 In the context of cancer therapy, recent studies revealed that a range of chemotherapeutic agents induce ICD, causing the conversion of the tumor microenvironment (TME) from inert to immunogenic by the release of DAMPs, leading to favorable therapeutic response.5,6 Hallmarks immunostimulatory DAMPs associated with chemotherapy-induced ICD include among others the exposure of calreticulin (CALR) on the plasma membrane, and release of adenosine triphosphate (ATP) and high mobility group box 1 protein (HMGB1).7 CALR is an endoplasmic reticulum (ER)-associated chaperone that translocate to the plasma membrane of dying cells in an eIF2A phosphorylation-dependent process, and acts as an “eat-me” signal for DCs at an early stage of ICD.8 Extracellular ATP released at an early stage of ICD acts as a “find me” signal, attracting and activating antigen presenting cells (APC) by binding purinergic receptors and promoting CD8+ T-cells priming.9 HMGB1 is a nuclear protein that is extracellularly released at a late stage of ICD and acts as “alarmin”, binding receptors such as TLR2, TLR4, and RAGE of immature DCs leading to DC maturation and cytotoxic T lymphocytes (CTLs) activation.10 The ability of some chemotherapeutic agents to induce immunogenic conditions in the TME has led to the development of combination therapeutic strategies of ICD-inducing chemotherapy and immune checkpoint blockers.11,12 Although these combination therapies have shown promising results for various cancer types, the effect of the therapy varies largely from patient to patient.13,14 Clinical evidence highlights that defects in immunogenic stress signaling adversely affect cancer prognosis and response to ICD-inducing therapies.15 These defects include poor cellular stress responses, low levels of DAMPs or their receptors, and genetic polymorphisms affecting pattern recognition receptor signaling.16 In vitro studies aiming at unrevealing the mechanism underlying DAMPs release associated with chemotherapy-induced ICD are carried out mostly using cell lines, which fail to recapitulate the patient-specific morphological and genetic features of the tumor.17 Therefore, the development of new methodological approaches to assess patient-specific differences while studying ICD in the context of cancer treatment might help the stratification of the patients and a better outcome prediction. In the past decades, personalized precision medicine has benefit from the advances made in 3D culture techniques using organoids.18,19 Organoids proved to be a more representative model of the in vivo situation compared to 2D cultures for a number of reasons, such as the presence of multiple organ-specific cell types, the ability to self-organize and to exert similar function of their tissue/organ counterparts.20 Furthermore, robust protocols are available to generate patient-derived organoids (PDOs) from many healthy and tumor tissue types.21 Thereby, we sought to extend the possibility to detect DAMPs associated with chemotherapy-induced ICD in PDOs. We exploited the colorectal cancer (CRC) PDOs collection generated in our previous work and applied oxaliplatin (OXA) and 5-fluorouracil (5FU), the two main components of the chemotherapy combination regimen FOLFOX broadly used for the treatment of colorectal cancer, to induce DAMPs release in PDOs.22–24 The aim of this study was to optimize a set of immunofluorescence and luminescence methods to detect the release of DAMPs in PDOs in response to chemotherapy agents. This study makes an important contribution to directing ICD research from the 2D cell monoculture system to the patient-derived 3D hetero-cell type system, paving the way for the development of patient-specific ICD investigation in vitro leading to better patient stratification.
Methods
Ethics Approval
PDOs used in this work were generated in a previous study using histologically verified primary colorectal tumor tissues obtained from adult-treated naïve patients undergoing surgical resection at the Medical University Hospital of Innsbruck with the approval of the medical ethical committee of the Medical University of Innsbruck, protocol AN2016-0194 366/4.9.22 Written informed consent for research was obtained from patients prior to tissue acquisition. This study complies with the Declaration of Helsinki.
PDOs Culture
PDOs were cultured in 30 µL droplets of 70% Geltrex (Thermo Scientific, Cat#A1413202) with PDO culture medium composed of Advanced DMEM/F12 (Thermo Scientific, Cat#12634028) supplemented with 10 mm HEPES solution (Sigma, Cat#H0887), 10 mL/L Penicillin-Streptomycin solution, 2 mm GlutaMAX (Thermo Scientific, Cat#3550061), 20% Rspondin conditioned medium, 10% Noggin conditioned medium, 20 mL/L B-27 supplement (Thermo Scientific, Cat#17504044), 1.25 mm N-Acetylcysteine (Sigma, Cat#A9165), 0.5 nM A83-01 (Tocris, Cat#2939), 10 mm SB202190 (Sigma, Cat#S7067), 50 ng/mL human EGF (Peprotech, Cat#AF-100-15), 100 mg/mL Primocin (Invivogen, Cat#ant-pm-2), and 10 mm Y27632 (AbMole, Cat#M1817). PDO culture medium was refreshed every two days.
Drug Sensitivity Assay
Organoids were enzymatically dissociated to single cells using 500 µL of TripLE (Sigma, Cat#T4174) for each culture droplet, incubated for 5 minutes at 37°C and reaction was stopped by adding an equal volume of Advanced DMEM/F12. Cells were filtered through a 70 µm strainer and centrifuged at 1500 rpm for 5 minutes. Cells were counted with a hemocytometer (Marienfeld Neubauer, Cat#0640010) and seeded at a number of 9000 cells/droplet for 1 day treatment, 8000 cells/droplet for 3 days treatment and 4500 cells/droplet for 6 days treatment into 96-black opaque-walled plates (Sarstedt GmbH, Cat#04.6000.024) in 7 µL droplets of 70% Geltrex covered with 100 µL PDO culture medium. After 3 days, culture medium was removed and replaced by 100 µL culture medium with 10 µM of Oxaliplatin (Biomol, Cat#AG-CR1-3592-M025) or 10 µM of 5FU (LKT Laboratories, Inc, Cat#F4480), or only culture medium for untreated control. Organoids viability was evaluated at day 1, day 3 and day 6 using a CellTiter-Glo 3D Cell viability assay (Promega, Cat#G9681) according to the manufacturer’s instructions. In brief, CellTiter-Glo 3D Reagent was thawed at 4° overnight. On the day of the measurement, the CellTiter-Glo 3D Reagent and the plate containing organoids were equilibrated at room temperature (22°) for 30 minutes. Culture medium was removed from the wells and 50 µL of fresh culture medium was added to each well together with 50 µL of the CellTiter-Glo 3D Reagent. The plate was shaken for 6 minutes at 500 rpm and incubated at room temperature for 25 minutes in the dark. Luminescence signal was recorded on the plate reader Tecan Infinite F200 Pro, i-control 2.0 Software.
Immunofluorescence Staining
For surface detection of Calreticulin and intracellular detection of HMGB1 in PDOs, we adapted the immunofluorescence protocol from Tesniere et al.25 For surface detection of Calreticulin, PDOs were seeded in 6 wells-plates (Sarstedt, Cat#83.3920) with 4 droplets of 30 µL of 70% Geltrex each well. After 3 days, culture medium was replaced with fresh medium containing 500 µM Oxaliplatin or 500 µM 5FU, or left untreated as control. After 4 hours incubation at 37°C and 5% CO2, 4 droplets were collected from the plate with a cell lifter (Corning, Cat#CLS3008), centrifuged for 5 minutes at 1500 rpm at 4°C and washed with cold Dulbecco’s Phosphate Buffered Saline (PBS) (Sigma, Cat#D8537). To remove the Geltrex, droplets were resuspended in 500 μL of Cell Recovery Solution (Corning, Cat#7340107P), cell suspension was incubated for 30 minutes at 4°C and organoids were washed 2 times with cold PBS by centrifuging 5 minutes at 1500 rpm at 4°C. Organoids were fixed with 0.25% ROTI Histofix Phosphate Buffered Formaldehyde (PFA) (Carl Roth, Cat#P087.4) in PBS for 5 minutes at 4°C, and washed 2 times with cold PBS. Organoids were stained for 45 minutes at 4°C in the dark with Anti-Calreticulin (Abcam, Cat#ab2907, lot.GR3248979-4) diluted 1:100 in Blocking Buffer (BB): PBS with 5% BSA (Sigma, #A6003), 10% goat serum (Sigma, Cat#G9023), 0.2% Tween (Sigma, Cat#P9416). Organoids were washed 5 times with Washing Buffer (WB): 0.3% fetal bovine serum (FBS) (Sigma, Cat#F7524) in PBS. Secondary antibody staining was performed with Goat Anti-Rabbit conjugated with Alexa Fluor 488 (1:250) (Abcam, Cat#ab150077, lot.GR3245102-1) in BB for 45 minutes at 4°C in the dark. Wheat Germ Agglutinin (WGA) conjugated with Alexa Fluor 633 (1:200) (Invitrogen, Cat#W21404 lot.2087692) was used for staining of plasma membranes. Organoids were washed twice with WB, fixed with 4% ROTI Histofix PFA for 20 minutes at room temperature and washed with WB for 2 times. Organoids were centrifuged 5 minutes at 1500 rpm at 4°C, supernatant was removed, organoids were resuspended in 20 μL Vectashield Vibrance Antifade Mounting Medium with DAPI (Vector Laboratories, Cat#H-2000-2 lot. ZJ0309), and transferred on SuperFrost Plus slides (VWR, Cat# 631-0108). Mounted organoids were covered with a Cover Glass (Corning, Cat#CLS285022) and left to dry overnight at room temperature.
For intracellular detection of HMGB1, PDOs were seeded as mentioned above for Calreticulin detection. After 3 days, culture medium was replaced with fresh medium containing 10 µM Oxaliplatin or 10 µM 5FU, or left untreated as control. After 48 hours incubation at 37°C and 5% CO2, 4 droplets were collected from the plate with a cell lifter, centrifuged for 5 minutes at 1500 rpm at 4°C and washed with cold PBS. Organoids were fixed with 4% ROTI Histofix PFA for 20 minutes at room temperature, washed 2 times with PBS, permeabilized with 0.1% Triton X-100 (Sigma, Cat#T9284) in PBS for 10 minutes at room temperature, and washed twice with PBS. PDOs were blocked with BB for 1 hour at room temperature and washed twice with PBS. Organoids were stained overnight at 4°C with Anti-HMGB1 (Cell Signaling, Cat. #3935S, lot.4) diluted 1:100 in BB. Organoids were washed 3 times in WB and secondary antibody staining was performed with Goat Anti-Rabbit conjugated with Alexa Fluor 488 (1:250) (Abcam, Cat#ab150077, lot.GR3245102-1) diluted in BB for 1 hour at room temperature in the dark. Wheat Germ Agglutinin (WGA) conjugated with Alexa Fluor 633 (1:200) (Invitrogen, Cat#W21404 lot.2087692) was used for staining of plasma membranes. Organoids were washed twice with WB and mounted as described above.
Organoids were imaged using a Zeiss LSM980 AiryScan2 Microscope (Carl Zeiss GmbH, Germany) with the Airyscan method, with a 63×1.40NA oil objective and images were acquired as three channel stacks. Acquired z-stacks were deconvolved to improve the final resolution with the Huygens Professional software (Scientific Volume Imaging, Hilversum, The Netherlands) with the Classic Maximum Likelihood Estimation (CMLE) algorithm, according to the manufacturer’s recommendations. The colocalization analysis was performed with the Colocalization Analyzer on the Huygens Professional between two channels of interest (Calreticulin/Plasma membrane or HMGB1/DAPI) using the calculated Gaussian Minimum threshold. The resulting overlap coefficients were analyzed by Prism software (v 10.4; GraphPad Software, Inc., La Jolla, CA, USA).
ATP Release Assay
Extracellular ATP release was detected using RealTime-Glo Extracellular ATP Assay (Promega, Cat#GA5010). Organoids were enzymatically dissociated to single cells and seeded at 9000 cells/well into 96-black opaque-walled plates (Sarstedt GmbH, Cat#04.6000.024) as described above. After 3 days, culture medium was replaced with 100 µL of fresh culture medium containing 1 mm of Oxaliplatin, or 1 mm 5FU or left untreated as control, and a 1× of RealTime-Glo Reagent. The plate was shaken at 500 rpm for 45 seconds and incubated for 4 hours at 37°C 5%CO2. Luminescence signal was recorded on the plate reader Tecan Infinite F200 Pro, i-control 2.0 Software.
Statistical Analysis
All assays were performed in triplicate. Data are presented as the mean ± standard error of the mean. Data were processed by Prism software (v 10.4; GraphPad Software, Inc., La Jolla, CA, USA). Differences in mean values between treated and control samples were computed by one-way ANOVA with post hoc Dunnet’s test. A value of P < 0.05 was considered statistically significant.
Results
Analysis of PDOs Sensitivity to OXA and 5FU Treatments
The combination of OXA and 5FU is the standard care for colorectal cancer.26 OXA is a platinum-based chemotherapeutic agent that acts as a strong ICD inducer, whereas the antimetabolite chemotherapeutic agent 5FU has been shown to only moderately induce ICD in CRC cell lines.25,27,28 To assess the effect of the drugs on the viability of organoids, 5 CRC PDOs generated and molecularly characterized in our previous work were selected (Table 1) and treated with OXA or 5FU.22 The response to the treatments over time varied for the different PDOs lines, with the most pronounced effect observed for CRC13 and CRC19 treated with OXA (Figure 1). Both CRC13 and CRC19 are classified as microsatellite instable (MSI), and their greater sensitivity to the treatment is consistent with previous reports showing that deficient mismatch repair (dMMR)/MSI CRC cell lines exhibit sensitivity to chemotherapeutics in vitro.29 Among the microsatellite stable (MSS) PDOs selected for this analysis, CRC17 shows the highest sensitivity to OXA. In our previous analysis we showed that CRC17 belongs to a small fraction of MSS tumors known to be transcriptionally more similar to the MSI-High tumors than to the MSS group, which agrees with the sensitivity to OXA observed.22,30 When treated with 5FU, MSI PDOs respond similarly to MSS (Figure 1), showing a less pronounced sensitivity than the one observed for OXA treatment, in line with prior studies demonstrating a lack of benefit of 5FU as adjuvant chemotherapy for dMMR/MSI CRC.31 The results of this assay indicate a heterogeneous response of different PDOs, particularly to OXA treatment.
 |
Table 1 Patients Clinical Information for the Corresponding CRC PDOs |
Analysis of CALR Exposure
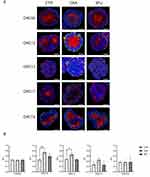
Exposure of CALR on the plasma membrane is the early phase hallmark of ICD. We optimized a protocol for immunofluorescence microscopy to analyze the translocation of CALR to the plasma membrane in PDOs treated with OXA and 5FU. We then quantified the exposure of CALR by calculating the colocalization of CALR and WGA, expressing it as overlap coefficient (OC). The results showed that OXA treatment induced an increment of CALR localized on the plasma membrane of CRC12 and CRC13 compared to the untreated control (Figure 2A and B). In contrast, none of the PDOs showed plasma membrane accumulation of CALR upon 5FU treatment.
Measurement of Extracellular ATP Release
ATP release from dying cells is one major hallmark of ICD. We refined an established luminescence assay to determine the extracellular ATP release from PDOs treated with OXA or 5FU compared to untreated PDOs. OXA treatment-induced secretion of ATP in CRC02, CRC12, CRC13 and CRC19. When PDOs were treated with 5FU, CRC19 showed a moderate increment in ATP secretion, while for the other PDOs the level of extracellular ATP appeared unaffected by the 5FU treatment (Figure 3).
Analysis of HMGB1 Release
The post-mortem release of HMGB1 is one typical feature of ICD. We optimized a protocol for immunofluorescence microscopy to analyze the translocation of HMGB1 from the nucleus to the cytoplasm in PDOs treated with OXA and 5FU. We then quantified the release of HMGB1 to the cytoplasm by calculating the colocalization of HMGB1 and DAPI, expressing it as overlap coefficient. The results showed that OXA treatment induced a decrement of HMGB1 localized in the nucleus of CRC12, CRC13 and CRC19 compared to the untreated control, while localization of HMGB1 was not altered in PDOs upon 5FU treatment (Figure 4A and B). These results are in line with the analysis of CALR exposure and ATP secretion (Figures 2 and 3) and indicate that 5FU does not trigger DAMPs in the CRC PDOs consistently, while OXA specifically triggers ICD-associated DAMPs in a subgroup of PDOs.
Discussion
Chemotherapy-induced ICD and DAMPs release relate to positive outcomes in cancer treatments. In vitro studies aimed at further understanding the role of ICD in cancer are mostly based on 2D cell lines that fail to recapitulate the tumor specificity of patients.25 Although studies based on 2D cultures have shed light on the mechanisms underlying ICD and the potential for induction of ICD by some chemotherapeutic agents on cancer cells, there is the strong need to move away from traditional 2D culture cancer cell lines embracing more complex 3D experimental systems.32 These systems have clinical relevance and reflect the phenotypic and genetic heterogeneity of primary tumors, allowing direct translation of in vitro findings to the clinic. The first step in this direction was made by developing 3D spheroids derived from tumor cell lines. Wernitznig and coworkers showed that spheroids derived from the colon carcinoma cell lines HCT-15, HCT-116 and HT-29 release the ICD-related DAMPs CALR, HMGB1 and ATP when treated with anticancer agents such as OXA, and the level of DAMPs release is drug-specific.33 However, even if spheroids offer an intermediate complexity between cancer cells grown in 2D monolayers and in vivo tumors, they lack the cell-type heterogeneity needed to fully recapitulate the tumor specificity of patients. In contrast, 3D PDOs retain intra- and inter-tumor heterogeneity and a previous study revealed a complexity of cell subtypes in OXA- and 5FU-resistant CRC samples by scRNA-seq that would not have been revealed by a simpler in vitro model.34
In our study, we optimized a series of assays to detect DAMPs release in a 3D organoid model. To set up the assays we exploit the CRC PDOs collection that we previously generated and characterized, we applied OXA and 5FU treatments to induce potential ICD and we optimized immunofluorescence and luminescence methods to detect the release of DAMPs. The response of the PDOs to the OXA and 5FU treatments was measured overtime using the CellTiter-Glo 3D Cell viability assay. In line with our expectations, with this approach we determined a heterogeneous response of the PDOs particularly to OXA treatment.
Exposure of CALR on the plasma membrane of 2D cell lines is typically determined by flow cytometry, immunofluorescence microscopy, or immunoblotting.19 Cytosolic translocation of HMGB1 in cell lines is also normally detected by immunofluorescence microscopy or immunoblotting. Flow cytometry and immunoblotting have a number of limitations when applied to organoid models. The main limiting step in flow cytometry is the need to disassociate the organoids into single cells suspension, a procedure that could introduce bias in the viability status of the cells unrelated to the treatment applied. In addition, organoids have heterogeneous size and morphology, and a standardized dissociation protocol could give different results on organoids cultures, introducing another level of bias in the results. Similarly, immunoblotting with organoid material presents some challenges: the heterogeneity in organoid size and morphology makes it difficult to apply a standardized cell lysis protocol, with results not comparable among different organoid cultures. Furthermore, studying protein translocation in immunoblot requires a crucial and delicate subcellular fractionation step to separate membrane, cytosolic and nuclear proteins. In addition, a substantial amount of organoids is required as starting material for this approach. Therefore, immunofluorescence microscopy seemed to be the most promising technique to be adapted for CALR and HMGB1 detection in organoid models. In our study, we successfully measured and quantified the intracellular translocation of CALR on the plasma membrane and HMGB1 in the cytoplasm of treated PDOs using an immunofluorescence microscopy approach. Upon OXA treatment, we could detect CALR translocation to the plasma membrane of CRC12 and CRC13, and cytoplasm translocation of HMGB1 in CRC12, CRC13 and CRC19. In contrast, 5FU treatment failed to promote CALR and HMGB1 intracellular translocation in all PDOs. With our approach, we were able to identify PDO-specific CALR and HMGB1 translocation following different treatments, and our results are in line with previous knowledge indicating OXA as a strong and 5FU as moderate ICD inducer.25,27,28
ICD-associated ATP secretion in cell lines can be assessed by monitoring the intracellular or the extracellular ATP content. Intracellular ATP is typically detected by staining with ATP-sensitive fluorochrome quinacrine and monitored by flow cytometry.9 As discussed earlier, flow cytometry is hardly applicable to the organoid model. Therefore, we choose to focus on the extracellular ATP secretion. We refined an established luminescence assay to determine ATP release in the culture medium of PDOs treated with OXA or 5FU. This rather straightforward approach allowed us to quantify changes in ATP secretion in different PDOs. In line with our expectations, we were able to detect an increment in ATP release in OXA-treated PDOs, and the amount of the secretion was PDO-dependent. Further, 5FU did not have an effect in ATP secretion in PDOs, with the exception of CRC19 that showed a moderate ATP secretion.
Overall, the results of this study indicate that among the PDOs analyzed, treatment with OXA triggers the release of the three major ICD-related DAMPs in CRC13 and CRC12. Of note, these two PDOs responded to OXA treatment differently in terms of cell viability over time, with CRC13 showing a much higher sensitivity than CRC12. This observation may indicate that induction of DAMPs release by chemotherapeutic agents may not be necessarily linked to a rapid reduction in cell viability in vitro. Based on the DAMPs release assays we performed, it can be hypothesized that both CRC13 and CRC12 are potential good responders to the combination of ICD-inducing chemotherapy and immune checkpoint blockers, underscoring the importance of developing experimental approaches that allow the recognition of patient-specific ICD activation in vitro, which could help predict patient response to therapies.
Our work has several limitations that can be addressed in future studies. One limitation of our study is the use of individual agents for treatments. In our study, we choose to focus on the effect of single chemotherapy agents to induce ICD in PDOs in order to better dissect the effect of each agent on different PDOs, taking advantage of the previous knowledge defining OXA as a strong ICD inducer and 5FU as moderate inducer. Several chemotherapeutic agents increase their effectiveness when administered in combination regiments, eg 5FU and OXA are normally administered together with folinic acid as combined chemotherapy regimen, named FOLFOX, to treat colorectal cancer.26 Therefore, further studies are required to explore the ICD-inducing potential of chemotherapeutic agents’ combination therapies in PDOs. Previous studies showed that CT-26 cells treated with FOLFOX presented an elevated CD47 and PD-L1 expression, and the combination of FOLFOX with CD47 and PD-L1 blocking antibodies increased survival and reduced tumor size in CT-26 tumor-bearing mice.35 The next steps in this line of research could exploit PDOs as models for ICD-studies, as proposed in our work, focusing further on combining pro-immunogenic drugs with anti-inhibitory antibodies, making the organoid model the bridge between pre-clinical studies and translation into the clinic. Another limitation of our study is the lack of the immune cell component in our 3D organoid in vitro system. The release of ICD-related DAMPs in tumor cells upon certain chemotherapy treatments results in the activation of dendritic cells and the subsequent antigen presentation to T cells that mediate tumor‐specific immune responses. In our experimental set-up, we were able to detect patient-specific DAMPs release in CRC organoids in response to chemotherapy treatments. To investigate the effect that DAMPs released by treated organoids might have on the immune system, further studies are needed to increase the complexity of the experimental set-up optimizing organoids and immune cells co-culture systems. Finally, with our study we intended to show a proof-of-concept that PDOs are suitable for personalized ICD studies, for which we used a relatively low number of PDOs lines. Future studies comprising a large cohort of PDOs are needed for further investigation to potentially identify patterns and biomarker for patient stratification. Despite the limitation discussed, to our knowledge, this is the first study employing CRC PDOs model for direct investigation of ICD, paving the way for further patient-specific studies essential to better translate basic preclinical research into novel treatments.
Conclusions
In this study, in vitro assays were optimized to evaluate ICD-related DAMPs release in CRC PDOs upon chemotherapy treatments. CALR translocation to the plasma membrane and HMGB1 translocation to the cytoplasm can be monitored in immunofluorescence followed by quantification of colocalization with subcellular markers. ATP secretion can be monitored by luminescence assay. This approach has the potential to identify ICD initiation in CRC PDOs in response to chemotherapy, and might be used to predict patients’ response to chemotherapy and immune checkpoint blockers combination therapy. In this work, we optimized the assays using CRC PDOs treated with OXA and 5FU, and we believe that the same approach could be adapted for the use in PDOs derived from other tissues and for testing different ICD-inducing drugs. We hope this approach will contribute to facilitating patient-specific ICD investigation in vitro and improve patient stratification.
Abbreviations
ICD, immunogenic cell death; DAMPs, damage associated molecular patterns; CRC, colorectal cancer; PDOs, patient-derived organoids; OXA, oxaliplatin; 5FU, 5-fluorouracil; CALR, calreticulin; HMGB1, high mobility group box 1; WGA, wheat germ agglutinin; ATP, adenosine triphosphate; RCD, regulated cell death; DCs, dendritic cell; TME, tumor microenvironment; ER, endoplasmic reticulum; APC, antigen presenting cells; CTLs, cytotoxic T lymphocytes; MSI, microsatellite instable; MSS, microsatellite stable; dMMR, deficient mismatch repair; OC, overlap coefficient; RLU, relative light units.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the European Research Council (project EPIC 786295 to Z.T.).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Casares N, Pequignot MO, Tesniere A, et al. Caspase-dependent immunogenicity of doxorubicin-induced tumor cell death. J Exp Med. 2005;202(12):1691–1701. doi:10.1084/jem.20050915
2. Galluzzi L, Vitale I, Aaronson SA, et al. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 2018;25(3):486–541. doi:10.1038/s41418-017-0012-4
3. Fucikova J, Kepp O, Kasikova L, et al. Detection of immunogenic cell death and its relevance for cancer therapy. Cell Death Dis. 2020;11(11):1013. doi:10.1038/s41419-020-03221-2
4. Kroemer G, Galassi C, Zitvogel L, Galluzzi L. Immunogenic cell stress and death. Nat Immunol. 2022;23(4):487–500. doi:10.1038/s41590-022-01132-2
5. Solari JIG, Filippi-Chiela E, Pilar ES, et al. Damage-associated molecular patterns (DAMPs) related to immunogenic cell death are differentially triggered by clinically relevant chemotherapeutics in lung adenocarcinoma cells. BMC Cancer. 2020;20(1):474. doi:10.1186/s12885-020-06964-5
6. Krysko DV, Garg AD, Kaczmarek A, Krysko O, Agostinis P, Vandenabeele P. Immunogenic cell death and DAMPs in cancer therapy. Nat Rev Cancer. 2012;12(12):860–875. doi:10.1038/nrc3380
7. Zhai J, Gu X, Liu Y, Hu Y, Jiang Y, Zhang Z. Chemotherapeutic and targeted drugs-induced immunogenic cell death in cancer models and antitumor therapy: an update review. Front Pharmacol. 2023;14:1152934. doi:10.3389/fphar.2023.1152934
8. Obeid M, Tesniere A, Ghiringhelli F, et al. Calreticulin exposure dictates the immunogenicity of cancer cell death. Nat Med. 2007;13(1):54–61. doi:10.1038/nm1523
9. Martins I, Wang Y, Michaud M, et al. Molecular mechanisms of ATP secretion during immunogenic cell death. Cell Death Differ. 2014;21(1):79–91. doi:10.1038/cdd.2013.75
10. Tesniere A, Panaretakis T, Kepp O, et al. Molecular characteristics of immunogenic cancer cell death. Cell Death Differ. 2008;15(1):3–12. doi:10.1038/sj.cdd.4402269
11. Wang Q, Ju X, Wang J, Fan Y, Ren M, Zhang H. Immunogenic cell death in anticancer chemotherapy and its impact on clinical studies. Cancer Lett. 2018;438:17–23. doi:10.1016/j.canlet.2018.08.028
12. Ouyang P, Wang L, Wu J, et al. Overcoming cold tumors: a combination strategy of immune checkpoint inhibitors. Front Immunol. 2024;15:1344272. doi:10.3389/fimmu.2024.1344272
13. Larroquette M, Domblides C, Lefort F, et al. Combining immune checkpoint inhibitors with chemotherapy in advanced solid tumours: a review. Eur J Cancer. 2021;158:47–62. doi:10.1016/j.ejca.2021.09.013
14. Pol J, Vacchelli E, Aranda F, et al. Trial watch: immunogenic cell death inducers for anticancer chemotherapy. Oncoimmunology. 2015;4(4):e1008866. doi:10.1080/2162402X.2015.1008866
15. Marei HE, Hasan A, Pozzoli G, Cenciarelli C. Cancer immunotherapy with immune checkpoint inhibitors (ICIs): potential, mechanisms of resistance, and strategies for reinvigorating T cell responsiveness when resistance is acquired. Cancer Cell Int. 2023;23(1):64. doi:10.1186/s12935-023-02902-0
16. Li D, Wu M. Pattern recognition receptors in health and diseases. Signal Transduct Target Ther. 2021;6(1):291. doi:10.1038/s41392-021-00687-0
17. Nayagom B, Amara I, Habiballah M, Amrouche F, Beaune P, de Waziers I. Immunogenic cell death in a combined synergic gene- and immune-therapy against cancer. Oncoimmunology. 2019;8(12):e1667743. doi:10.1080/2162402X.2019.1667743
18. Bose S, Clevers H, Shen X. Promises and challenges of organoid-guided precision medicine. Med. 2021;2(9):1011–1026. doi:10.1016/j.medj.2021.08.005
19. Galluzzi L, Vitale I, Warren S, et al. Consensus guidelines for the definition, detection and interpretation of immunogenic cell death. J Immunother Cancer. 2020;8(1). doi:10.1136/jitc-2019-000337
20. Kim J, Koo BK, Knoblich JA. Human organoids: model systems for human biology and medicine. Nat Rev Mol Cell Biol. 2020;21(10):571–584. doi:10.1038/s41580-020-0259-3
21. Driehuis E, Kretzschmar K, Clevers H. Establishment of patient-derived cancer organoids for drug-screening applications. Nat Protoc. 2020;15(10):3380–3409. doi:10.1038/s41596-020-0379-4
22. Plattner C, Lamberti G, Blattmann P, et al. Functional and spatial proteomics profiling reveals intra- and intercellular signaling crosstalk in colorectal cancer. iScience. 2023;26(12):108399. doi:10.1016/j.isci.2023.108399
23. Gustavsson B, Carlsson G, Machover D, et al. A review of the evolution of systemic chemotherapy in the management of colorectal cancer. Clin Colorectal Cancer. 2015;14(1):1–10. doi:10.1016/j.clcc.2014.11.002
24. Qu F, Wu S, Yu W. Progress of immune checkpoint inhibitors therapy for pMMR/MSS metastatic colorectal cancer. Onco Targets Ther. 2024;17:1223–1253. doi:10.2147/OTT.S500281
25. Tesniere A, Schlemmer F, Boige V, et al. Immunogenic death of colon cancer cells treated with oxaliplatin. Oncogene. 2010;29(4):482–491. doi:10.1038/onc.2009.356
26. McQuade RM, Stojanovska V, Bornstein JC, Nurgali K. Colorectal cancer chemotherapy: the evolution of treatment and new approaches. Curr Med Chem. 2017;24(15):1537–1557. doi:10.2174/0929867324666170111152436
27. Perego P, Robert J. Oxaliplatin in the era of personalized medicine: from mechanistic studies to clinical efficacy. Cancer Chemother Pharmacol. 2016;77(1):5–18. doi:10.1007/s00280-015-2901-x
28. Liang YH, Tsai JH, Cheng YM, et al. Chemotherapy agents stimulate dendritic cells against human colon cancer cells through upregulation of the transporter associated with antigen processing. Sci Rep. 2021;11(1):9080. doi:10.1038/s41598-021-88648-z
29. Ye T, Lin A, Qiu Z, et al. Microsatellite instability states serve as predictive biomarkers for tumors chemotherapy sensitivity. iScience. 2023;26(7):107045. doi:10.1016/j.isci.2023.107045
30. Joanito I, Wirapati P, Zhao N, et al. Single-cell and bulk transcriptome sequencing identifies two epithelial tumor cell states and refines the consensus molecular classification of colorectal cancer. Nat Genet. 2022;54(7):963–975. doi:10.1038/s41588-022-01100-4
31. Kawakami H, Zaanan A, Sinicrope FA. Microsatellite instability testing and its role in the management of colorectal cancer. Curr Treat Options Oncol. 2015;16(7):30. doi:10.1007/s11864-015-0348-2
32. Sun L, Wan AH, Yan S, et al. A multidimensional platform of patient-derived tumors identifies drug susceptibilities for clinical lenvatinib resistance. Acta Pharm Sin B. 2024;14(1):223–240. doi:10.1016/j.apsb.2023.09.015
33. Wernitznig D, Kiakos K, Del Favero G, et al. First-in-class ruthenium anticancer drug (KP1339/IT-139) induces an immunogenic cell death signature in colorectal spheroids in vitro. Metallomics. 2019;11(6):1044–1048. doi:10.1039/c9mt00051h
34. Chen G, Gong T, Wang Z, et al. Colorectal cancer organoid models uncover oxaliplatin-resistant mechanisms at single cell resolution. Cell Oncol. 2022;45(6):1155–1167. doi:10.1007/s13402-022-00705-5
35. Alimohammadi R, Mahmoodi Chalbatani G, Alimohammadi M, et al. Dual blockage of both PD-L1 and CD47 enhances the therapeutic effect of oxaliplatin and FOLFOX in CT-26 mice tumor model. Sci Rep. 2023;13(1):2472. doi:10.1038/s41598-023-29363-9
 © 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2025 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 4.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.