Back to Journals » Journal of Inflammation Research » Volume 17
The Mechanism by Which Cyperus rotundus Ameliorates Osteoarthritis: A Work Based on Network Pharmacology
Authors Du MD, He KY, Fan SQ, Li JY, Liu JF, Lei ZQ, Qin G
Received 6 August 2024
Accepted for publication 26 September 2024
Published 29 October 2024 Volume 2024:17 Pages 7893—7912
DOI https://doi.org/10.2147/JIR.S483652
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tara Strutt
Min-Dong Du,1,2,* Kai-Yi He,3,* Si-Qi Fan,3 Jin-Yi Li,3 Jin-Fu Liu,3 Zi-Qiang Lei,3 Gang Qin3
1Department of Osteoarthrosis, Xing-An Jieshou Orthopedics Hospital, Guilin, People’s Republic of China; 2Department of Orthopaedic Trauma and Hand Surgery, The First Affiliated Hospital of Guangxi Medical University, Nanning, People’s Republic of China; 3Department of Osteoarthrosis, The First Affiliated Hospital of Guangxi Traditional Chinese Medical University, Nanning, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Gang Qin, Department of Osteoarthrosis, The First Affiliated Hospital of Guangxi Traditional Chinese Medical University, 89-9 Dongge Road, Nanning, Guangxi, 530023, People’s Republic of China, Tel +86-771-5361264, Email [email protected]
Background: Cyperus rotundus (CR) is widely used in traditional Chinese medicine to prevent and treat a variety of diseases. However, its functions and mechanism of action in osteoarthritis (OA) has not been elucidated. Here, a comprehensive strategy combining network pharmacology, molecular docking, molecular dynamics simulation and in vitro experiments was used to address this issue.
Methods: The bioactive ingredients of CR were screened in TCMSP database, and the potential targets of these ingredients were obtained through Swiss Target Prediction database. Genes in OA pathogenesis were collected through GeneCards, OMIM and DisGeNET databases. Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were performed using DAVID database. STRING database and Cytoscape 3.10 software were used to construct “component-target-pathway” network, and predict the core targets affected by CR. The binding affinity between bioactive components and the core targets was evaluated by molecular docking and molecular dynamics simulation. The therapeutic activity of kaempferol on chondrocytes in inflammatory conditions was verified by in vitro experiments.
Results: Fifteen CR bioactive ingredients were obtained, targeting 192 OA-related genes. A series of biological processes, cell components, molecular functions and pathways were predicted to be modulated by CR components. The core targets of CR in OA treatment were AKT serine/threonine kinase 1 (AKT1), interleukin 1 beta (IL1B), SRC proto-oncogene, non-receptor tyrosine kinase (SRC), BCL2 apoptosis regulator (BCL2), signal transducer and activator of transcription 3 (STAT3), epidermal growth factor receptor (EGFR), hypoxia-inducible factor 1 subunit alpha (HIF1A), matrix metallopeptidase 9 (MMP9), estrogen receptor 1 (ESR1) and PPARG orthologs from vertebrates (PPARG), and the main bioactive ingredients of CR showed good binding affinity with these targets. In addition, kaempferol, one of the CR bioactive components, weakens the effects of IL-1β on the viability, apoptosis and inflammation of chondrocytes.
Conclusion: Theoretically, CR has great potential to ameliorate the symptoms and progression of OA, via multiple components, multiple targets, and multiple downstream pathways.
Keywords: osteoarthritis, Cyperus rotundus, network pharmacology, molecular docking
Graphical Abstract:

Introduction
Osteoarthritis (OA) is a common degenerative joint disease, mainly characterized by articular cartilage injury, subchondral osteosclerosis and synovial tissue inflammation.1,2 Aging, joint trauma and obesity are important risk factors for the pathogenesis of OA.3 In recent years, the prevalence of OA has gradually increased.4,5 Epidemiological surveys show that OA affects about 260 million people worldwide,6 accounting for 2.4% of the total number of people with disabilities.3,5 At present, the clinical treatment of OA mainly uses non-steroidal anti-inflammatory drugs or intra-articular local injection of glucocorticoid to reduce symptoms and restore joint function.7 However, long-term use of these drugs leads to side effects associated with gastrointestinal, kidney, and cardiovascular symptoms.8 Therefore, there is an urgent need to develop effective treatments with better bioavailability and less systemic toxicity.
It is important to note that the pathogenesis of human disease is often associated with abnormalities in multiple genes, proteins and pathways. Specifically, in OA, multiple pathways related to chondrocyte injury, inflammatory response, and extracellular matrix synthesis are dysregulated, and these pathways have synergistic effects in the occurrence and progression of OA, and also cause the diversification of clinical symptoms.9,10 Chinese herbal medicine has “multi-component” and “multi-target” anti-disease mechanism, which reflects a certain clinical application prospect. Herbal medicine has been widely used to treat a range of diseases, including OA, in some developing countries.11,12 In Traditional Chinese Medicine (TCM) literature Compendium of Materia Medica, Cyperus rotundus (CR) is used to treat Han Ning Qi Zhi (qi stagnation due to cold congealing), which is a common cause of limb pain. Modern medical research has reported that CR has the pharmacological effects of ameliorating depression, regulating menstrual disorder and relieving pain.13 CR contains a variety of bioactive substances, including alkaloids, flavonoids, polyphenols and terpenoids.13–15 Modern pharmacological studies have shown that CR has anti-oxidation, anti-depression, anti-arthritis, anti-tumor, hypoglycemic and anti-bacterial effects, and is often used to treat diseases of nervous system, cardiovascular system and digestive system in China.16–18 Notably, the anti-inflammatory effects of CR have been widely reported.16 Additionally, α-Cyperone, one of the main bioactive components of CR, can ameliorate OA in mice by inhibiting NF-κB and MAPK signaling pathways, reducing chondrocyte inflammation and extracellular matrix degradation.19 These studies suggest that CR is promising to treat OA as a complementary option for the current treatment strategies, which is consistent with the theory of TCM and has been supported by existing pharmacological studies.13–18
Network pharmacology is a new research approach to elucidate the underlying mechanisms of a drug on specific diseases through bioinformatics analysis and target prediction.20 This study aims to clarify the mechanism of CR in the treatment of OA with network pharmacology. First of all, the potential bioactive components of CR were summarized. Secondly, their respective targets and OA-related genes were collected, and cross-over analysis was performed. Next, based on the potential CR targets in OA treatment, bioinformatics analysis and topological analysis were performed, the further identify the crucial targets and pathways affected by CR. In addition, in vitro experiments were performed to verify the anti-OA effect of kaempferol, one of CR bioactive components.
Materials and Methods
Bioactive Ingredients of CR and Target Prediction
Traditional Chinese Medicine System Pharmacology Database (TCMSP; https://www.tcmsp-e.com/) was searched to obtain the components of CR, and oral bioavailability (OB)≥30% and drug-like (DL)≥0.18 were used as screening criteria.21 PubChem (https://pubchem.ncbi.nlm.nih.gov/)22 was searched for chemical structure of the components with SMILES format, and SwissTargetPrediction database (http://www.swisstargetprediction.ch/) was searched to predict the targets of CR components. The target with “Probability” >0.01 was set as the drug target.23,24 Gene names and gene IDs were confirmed by UniProt (https://www.uniprot.org/).
Crucial Genes in OA Pathogenesis
With “osteoarthritis” as the key word, via online human Mendel database (OMIM, https://omim.org/,25 human genome database (GeneCards, https://www.genecards.org/)26 and human disease gene database (DisGeNET, https://www.disgenet.org/)27 were searched for the crucial genes involved in OA pathogenesis.
Network Construction of Potential Targets in OA Treatment with CR
Draw Venn Diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/) was used to get the gene targets in the intersection. These targets were defined as potential targets for OA with CR treatment. Bioactive components and disease targets were introduced into Cytoscape 3.10 software to construct the network diagram, and the core components of CR were screened according to the degree value.
Gene Enrichment Analysis
DAVID database (https://david.ncifcrf.gov/tools.jsp)28 was used for gene ontology (GO) enrichment analysis, including biological process (BP), cells (CC), molecular function (MF), and Kyoto Encyclopedia of Enrichment of Genes and Genomes (KEGG) enrichment analysis, based on the genes in the intersection of CR targets, and OA therapeutic targets (in other words, CR’s targets in OA treatment). With P < 0.01 as the filter, key biological functions and affected signal pathways in OA treatment with CR were predicted.
Construction and Analysis of Protein–Protein Interaction (PPI) Network
The target of CR for OA treatment was introduced into the “Multiple Proteins” in STRING database (https://cn.string-db.org/),29 and the ‘organism’ was specified as “Homo sapiens”, and the lowest interaction score was set to medium confidence 0.4. After removing the free points, the PPI network diagram was obtained, and the TSV file was downloaded and saved. TSV files were imported into Cytoscape 3.10 software for visualization, using the Centiscape 2.2 plugin to screen the core targets for CR treatment of OA.
Molecular Docking
AutoDock Vina (version 1.1.2) and PyMOL 2.4.0 software30 were used for molecular docking. The X-ray crystal structures of the selected proteins were obtained from the Protein Data Bank (PDB) database (https://www.rcsb.org/).31 Then, the PDB file is opened with PyMOL software, water molecules and heteroatoms are removed from the protein crystal structure, hydrogen atoms are added, and the charge is calculated. From PubChem database (https://pubchem.ncbi.nlm.nih.gov/), 3D chemical structures of the bioactive ingredients of CR were downloaded, and the files were saved as SDF format, and transformed with OpenBabel software (version 3.1.1)32 into mol2 format. Next, the 3D structure of the biocomponent and the targets were converted into a file in PDBQT format via AutoDockTools (version 1.5.7).33 Finally, AutoDock Vina software was used for molecular docking, and PyMOL software was used to visualize the results. The binding energy was calculated to evaluate the binding affinity between the components and the targets.
Molecular Dynamics Simulation
The general AMBER force field (gaff) was used for small molecule, and the ff14SB force field was used for the protein. The default protonation state in Amber14 was used for amino acid residues, which were hydrotreated with the tleap module. The Gaussian 09 software was used to optimize the structure of the protein, and the electrostatic potential was calculated based on B3LYP/6-31G. The charge of the small molecule was obtained by electrostatic potential fitting by Antechamber module using RESP method. The molecular mechanics optimization and molecular dynamics simulation of the complex was performed using the sander or pmemd program in Amber14. After the complex was added with counterbalance ions to maintain the neutrality of the system, the whole system was placed in the cuboid water box of TIP3P, and the water box extended to the three-dimensional space of the solute molecule 10Å × 10Å × 10Å. A two-step energy optimization operation was performed before molecular dynamics simulation. First, the energy optimization of solute molecular water, including 2500 steps of steepest descent method and 2500 steps of conjugate gradient method. Then, the energy optimization of the whole system is carried out for 5000 steps, including 2500 steps of steepest descent method and 2500 steps of conjugate gradient method. Particle mesh Ewald (PME) method was used to deal with the long range electrostatic interaction, and the SHAKE method was used to constrain all the bonds connected with hydrogen atoms, and the time step was set to 2 fs. A cut-off value of 10Å was used for non-bond interactions. The whole system was heated from 0 K to 300 K in 60 ps using constraints at constant volume, then the solvent density was equalized in a thermostatic pressure system (T = 300 K, P = 1 atm), and finally 100 ns was sampled at constant pressure and one frame (conformation) per ps was saved for subsequent analysis.
Cell Culture
Human chondrocytes C28/I2 were purchased from Kebai (Nanjing, China). The cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Rockville, MD, USA), 100 U/mL penicillin and 100 μg/mL streptomycin (Gibco, Rockville, MD, USA), in a humidified incubator at 37°C in 5% CO2.
Interleukin-1β (IL-1β) and Kaempferol Treatment
To mimic inflammatory condition in OA pathogenesis, C28/I2 cells were inoculated with a density of 5×105 cells/wells in a 6-well plate and exposed to 2 mL medium (containing 1% FBS) containing 10 ng/mL IL-1β (Beyotime, Shanghai, China).34,35 To study the effects of kaempferol (Sigma-Aldrich, St. Louis, MO, USA) on chondrocytes, C-28/I2 cells were treated with kaempferol at different concentrations (0, 10, 25, 50, 80, and 100 μM) for 24 h. To investigate the effect of kaempferol on IL-1β-induced chondrocytes, the cells were treated with 10 ng/mL of IL-1β and 50 μM or 100 μM of kaempferol for 24 h. Untreated cells served as the controls.
Cell Viability Assay
The cell viability was determined by 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) method. In short, C28/I2 cells were inoculated at a density of 5×103 cells/well and incubated overnight in a 96-well plate. Then, 20 μL of MTT reagent (5 mg/mL, Sigma, St. Louis, MO, USA) was added to each well and the cells were incubated at 37 °C for 4 h. Subsequently, 150 μL dimethyl sulfoxide (DMSO, Sigma, St. Louis, MO, USA) was added to each well, and the plate was shaken for 15 min to dissolve the crystals. Finally, the cell viability was measured at 450 nm wavelength using a microplate reader (Dynatech Labs, Chantilly, VA, USA).
Evaluation of Caspase-3 Activity
According to the manufacturer’s protocol, a colorimetric kit (Beyotime, Shanghai, China) was used to detect the activity of caspase-3 by colorimetry. In short, the cells were washed twice with cold phosphate buffer saline (PBS) and cleaved for 10 min in lysis buffer on ice. The cell lysate was centrifuged at 14,000× g for 10 min. The supernatant was collected and supplemented with 5 μL of caspase substrate to detect caspase-3 activity and incubated in a 96-well plate in a 37°C incubator for 4 h. Finally, the absorbance value was read at 405 nm wavelength using a microplate reader (Dynatech Labs, Chantilly, VA, USA).
Flow Cytometry
An Annexin V-Fluorescein Isothiocyanate (FITC)/Propidium Iodide (PI) apoptosis detection kit (Beyotime, Shanghai, China) was used to detect the apoptosis of chondrocytes. In short, C-28/I2 cells were inoculated in a 6-well plate with a density of 4×105 cells/well and incubated overnight at 37°C. The cells were collected and washed twice with PBS. The cells were then labeled in the dark with the dye in 500 μL 1× binding buffer containing 10 μL PI and 5 μL Annexin V-FITC for 5 min. Finally, after the cells were gently washed with the binding buffer, the apoptosis rate of the chondrocytes was measured by a flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
C28/I2 chondrocytes were collected, and total RNA was extracted using a TRIzol kit (Invitrogen, Carlsbad, CA, USA). cDNA was synthesized from 1 μg of total RNA using a M-MLV reverse transcriptase kit (Invitrogen, Carlsbad, CA, USA) in a final volume of 20 μL. qRT-PCR was performed using ABI 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA), and the cDNA was amplified with a SYBR Premix Ex Taq kit (Takara, Otsu, Japan). The primer sequences are as follows: inducible nitric oxide synthase (iNOS) forward primer, 5’-GGCAGCCTGTGAGACCTTG-3’, reverse primer, Inducible nitric oxide Synthase (INOS) forward primer, 5’- GGCAGCCTGTGAGACCTTG-3‘, reverse primer, 5’-GCATTGGAAGGAAGGAGCGTTTC-3’; Epoxide 2 (COX2) positive, 5’-TGCTGGTGGAAAAACCTCGT-3’, reverse primer, 5’-AAAACCCACTTCGCCTCCAA-3’; tumor necrosis factor-α (TNF-α) positive, 5’ -GCGTGTTCATCCGTTCTCTAC-3‘, reverse primer, 5’ -TACTTCAGCGTCTCGTGTGTTCT-3‘; interleukin-6 (IL-6) positive, 5’-CACTCACCTCTTCAGAACGAAT-3’, reverse primer, 5’-GCTGCTTTCACACATGTTACTC-3’, and β-actin positive, 5’-CCTGGCACCCAGCACAAT-3’, reverse primer, 5’-GGGCCGGACTCGTCATCG-3’.
Statistical Analysis
All experiments were conducted independently in triplicate. All data were expressed as “mean ± standard deviation (SD)”. SPSS 21.0 software (IBM Corp., Armonk, NY, USA) was used for statistical analyses of the data. Comparisons between groups were made using the Student’s t test or one-way ANOVA with Tukey’s post hoc tests. P < 0.05 was considered statistically significant.
Results
Screening of CR Bioactive Ingredient Targets and OA Regulators
According to the screening criteria of OB ≥ 30% and DL ≥ 0.18, 15 CR bioactive ingredients were obtained from the TCMSP database, as shown in Table 1. The chemical structures of these bioactive ingredients are shown (Figure 1). Subsequently, the Swiss Target Prediction database was used to predict the possible targets of CR components, and 407 potential targets were finally obtained. In addition, 3158, 30 and 331 OA-related genes were retrieved with the keyword “osteoarthritis” from GeneCards, OMIM and DisGeNET databases, respectively. After merging and removing the duplicate targets, a total of 3251 OA-related genes were obtained.
 |
Table 1 Active Components of Cyperus Rotundus |
 |
Figure 1 Chemical structure of 15 CR bioactive ingredients. |
Target Screening and Network Analysis for CR Treatment of OA
The targets of CR components were intersected with the genes in OA pathogenesis, and a total of 192 target genes were obtained (Figure 2A). Then, 192 targets were imported into Cytoscape 3.10 software to construct the “drug-bioactive ingredient-target” network to further show the relationship among CR bioactive ingredients and OA-related genes. The network consisted of 210 nodes and 795 edges (Figure 2B). The greater the degree value, the greater the possibility of the compound playing a therapeutic role. The top 10 components, with the highest degree values, were sugeonyl acetate, rosenonolactone, luteolin, kaempferol, chryseriol, quercetin, isorhamnetin, 8-Prenylkaempferol, tetrahydropalmatine and leucocianidol (Figure 2C). Therefore, we supposed that the above 10 compounds might be the important components of CR to ameliorate OA progression.
GO and KEGG Analyses of the Potential Targets of CR in OA Treatment
In order to further clarify the mechanism of CR in the treatment of OA, GO analysis and KEGG enrichment analysis were performed based on 192 genes. A total of 514 GO terms were obtained (P<0.01), including 356 biological process (BP) terms, 58 cellular component (CC) terms and 100 molecular function (MF) terms. The top 10 BP, CC, and MF terms were selected for visualization (Figure 3A). According to BP’s analyses, the functions of CR bioactive ingredients in treating OA were mainly associated with protein phosphorylation, positive regulation of cell migration and positive regulation of MAP kinase activity and response to xenobiotic stimulus, etc. (Figure 3A). The potential targets of CR in the treatment of OA were also associated with receptor complex, cytoplasm, plasma membrane, cytosol and macromolecular complex (Figure 3A). Protein serine/threonine/tyrosine kinase activity, protein kinase activity, protein tyrosine kinase activity, enzyme binding and RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding were also significantly associated with CR (Figure 3A). In addition, KEGG enrichment analysis showed that the potential targets of CR in OA treatment were enriched in 134 related pathways (P<0.01). The top 30 KEGG pathways were visualized. As shown, target genes of CR therapy for OA were mainly involved in pathways in cancer (hsa05200), EGFR tyrosine kinase inhibitor resistance (hsa01521), proteoglycans in cancer (hsa05205), PI3K-Akt signaling pathway (hsa04151), AGE-RAGE signaling pathway in diabetic complications (hsa04933), prostate cancer (hsa05215), Lipid and atherosclerosis (hsa05417), chemical carcinogenes-receptor activation (hsa05207) and focal adhesion (hsa04510) pathways (Figure 3B). In addition, we grouped the top 30 enriched KEGG pathways. The results showed that these KEGG pathways were mainly divided into four categories, including environmental information processing, cellular processes, organismal systems, and human diseases (Figure 3C). Additionally, the targets in the PI3K-Akt pathway were colored using KEGG PathView with red, which were the potential targets of CR in the treatment of OA (Figure 3D).
Construction of “Component-Target-Pathway” Network Diagram
Next, the targets on the top 10 signaling pathways and the targets of CR components were imported into Cytoscape 3.10 software to construct the “component-target-pathway” network diagram. As shown, the 15 bioactive ingredients of CR may play a key role in the top 10 pathways associated with OA by acting on 193 targets (Figure 4).
 |
Figure 4 “Component-target-pathway” network diagram established by Cytoscape 3.10 software. |
PPI Network Construction
Subsequently, the 192 gene targets of CR in the treatment of OA were imported into STRING database to construct PPI network. The network contained 191 nodes and 2976 edges (Figure 5A). The nodes represented proteins and the edges represented protein–protein interactions. Cytoscape 3.10 software was used for PPI network analysis. All targets were arranged in circles according to the degree value, and the larger the degree value, the darker the color, and the more important it was in the PPI network (Figure 5B). As shown, inflammation-related proteins (STAT3, SRC, IL-1β, TLR4, GSK3β), extracellular matrix degradation-related proteins (MMP2, MMP9) showed remarkable importance, which suggested that CR could probably modulate inflammatory response and cartilage degradation to ameliorate OA. In addition, core cluster proteins were analyzed using the Centiscape 2.2 plug-in in Cytoscape 3.10 software. With betweenness centrality ≥189.57 (Figure 6A), closeness centrality ≥0.003 (Figure 6B) and degree ≥31.16 (Figure 6C) as the screening criteria, 41 core genes were obtained (Figure 6D). The top 10 core gene targets are AKT1, IL1B, SRC, BCL2, STAT3, EGFR, HIF1A, MMP9, ESR1, and PPARG (Table 2).
 |
Table 2 Summary of the Top 10 Core Targets |
 |
Figure 6 Screening of core targets in PPI network. (A–C) Screening flowchart of Centiscape 2.2 plug-in. (D) Core targets of PPI network nodes. The larger the degree value, the larger the font. |
Molecular Docking and Molecular Dynamics Simulation
In order to further explore the downstream mechanism of CR in the treatment of OA, AutoDock Vina software was used for molecular docking. Six bioactive ingredients with the highest degree values (sugeonyl acetate, rosenonolactone, luteolin, kaempferol, chryseriol, quercetin) were docked with six representative core proteins in PPI network (AKT1, IL1B, SRC, BCL2, STAT3 and EGFR). Binding affinity less than −4.25 kcal/mol indicates that two molecules have standard binding ability; less than −5.0 kcal/mol indicates good binding; and less than −7.0 kcal/mol indicates strong binding activity.36 The results of binding affinity of key bioactive components with core proteins are shown in Table 3. It showed that sugeonyl acetate, rosenonolactone, luteolin, kaempferol, chryseriol, and quercetin could, respectively, bind with AKT1 (PDB ID: 8Q61) and IL1B (PDB ID: 8Q61), respectively. 31BI), SRC (PDB ID: 4M4Z), BCL2 (PDB ID: 5UUK), STAT3 (PDB ID: 6NUQ) and EGFR (PDB ID: 5Y9T), with good or strong binding affinity (Table 4 and Figure 7). The data mentioned above implied that the bioactive component of CR could ameliorate OA pathogenesis via binding with these proteins, most of which were involved in inflammation and cell apoptosis. Kaempferol has been reported to protect chondrocytes,37 so kaempferol was selected for further investigation. AKT1 had the highest docking score with it, so molecular dynamics simulation was performed based on kaempferol and AKT1. As shown, kaempferol-AKT1 complex was in a stable state at about 5ns, indicating that their intersection was stable, and the root mean square deviation (RMSD) trajectory of ligand (kaempferol) and active pocket of AKT1 fluctuated very little and maintained a stable state on the whole (Figure 8A). In order to explain the conformational fluctuations and local dynamics in the binding state of protein and compound, the average fluctuations of each amino acid residue were calculated, and then the root mean square fluctuation (RMSF) values of amino acid residues in the protein were investigated. As shown (Figure 8B), the RMSF value of the amino acid residues of AKT1 protein did not fluctuate as a whole, but only in the cavity where small molecules were located, which further supported that the binding between kaempferol and AKT1 was stable. Next, the binding free energy was calculated using MM/PBSA based on the conformation obtained by molecular dynamics simulation. It was found that the main beneficial contribution of small molecule binding is van der Waals force (ΔEvdw). It can also be seen from the connection diagram that small molecules are in the hydrophobic cavity of proteins, forming a strong van der Waals force. The second is the electrostatic interaction force (ΔEele). It was worth noting that the total binding free energy (ΔGTot) of protein binding is −27.37 kcal/mol, indicating that kaempferol has strong-binding ability to AKT1 (Figure 8C). Next, energy decomposition was performed. The main amino acids were Trp80 (−1.00 kcal/mol), Glu200 (−1.27 kcal/mol), Leu212 (−0.64 kcal/mol), Val271 (−0.84 kcal/mol) and Ile291 (−1.19kcal/mol) and Thr292 (−1.59 kcal/mol) (Figure 8D). This result was consistent with the result of molecular docking, and the binding energy with amino acids Glu200, Ile291 and Thr292 was stronger, and amino acids adjacent to small molecules also produce strong interaction. Additionally, in order to better compare the molecular dynamics process of kaempferol and AKT1 protein, a conformation was selected every 20ns to observe and compare the movement trajectory. As shown (Figure 8E), small molecules fluctuated slightly at the beginning stage, and then gradually turned to a stable state, and the movement of small molecules after binding was not large, indicating the binding was stable.
 |
Table 3 Binding Energy Between the Active Components of Cyperus Rotundus and the Key Targets in the PPI Network |
 |
Table 4 Docking Center and Binding Pocket of Compounds and Targets |
 |
Figure 7 Molecular docking of key components of CR and core targets of OA. Light blue indicates amino acid residues around the binding bag, and dark blue indicates the bioactive ingredient. |
Effects of Kaempferol on the Viability, Apoptosis and Inflammatory Response of Chondrocyte Treated with IL-1β
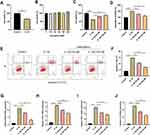
In order to verify the protective role of CR for chondrocytes, we selected kaempferol, a key component of CR, to perform in vitro experiments. C28/I2 cells were treated with 10 ng/mL IL-1β for 2 h to establish an in vitro model of OA. IL-1β significantly reduced the viability of C-28/I2 cells compared to the control group (Figure 9A). Subsequently, the cytotoxicity of kaempferol to chondrocytes was evaluated at different concentrations (0, 10, 25, 50, 80, and 100 μM). The results showed that different concentrations of kaempferol did not produce significant cytotoxicity to C-28/I2 cells (Figure 9B). Next, 50 μM or 100 μM kaempferol as a medium dose or high dose, respectively, to treat chondrocytes. As shown, kaempferol attenuated the injury and caspase-3 activation of C-28/I2 cell viability induced by IL-1β in a dose-dependent manner (Figure 9C and D). In addition, flow cytometry showed that IL-1β treatment significantly increased the apoptosis of C-28/I2 cells compared with the control group, and kaempferol inhibited IL-1β-induced apoptosis of C-28/I2 cells in a dose-dependent manner (Figure 9E and F). We also examined the effects of kaempferol on the expression of inflammatory cytokines (INOS, COX2, TNF-α, and IL-6) in C-28/I2 cells. The results showed that kaempferol reversed the upregulation of INOS, COX2, TNF-α, and IL-6 in chondrocytes induced by IL-1β in a dose-dependent manner (Figure 9G–J). Overall, these results suggested that kaempferol can reduce IL-1β-induced chondrocyte injury by promoting cell viability and inhibiting apoptosis and inflammatory responses.
Discussion
OA is a common degenerative joint disease whose incidence increases with age, and had significant adverse impact on the patients’ health and quality of life.38 Traditional Chinese medicine has multi-target and multi-pathway biological effects and has been applied to treat OA as adjunctive therapy.39,40 CR is widely used in traditional Chinese medicine to prevent/treat a variety of diseases.41–43 In the present work, our data showed that CR contained at least 15 bioactive ingredients, targeting 407 genes, of which 192 genes were associated with OA pathogenesis. It was found that the components in CR, such as sugeonyl acetate, rosenonolactone, luteolin, kaempferol, chryseriol and quercetin, had high regulatory potential on the proteins of OA pathogenesis by constructing a “component-target-pathway” network. Some previous studies have reported the role of these components on ameliorating inflammation of chondrocytes or blocking OA pathogenesis. For instance, luteolin inhibits IL-1β-induced chondrocyte inflammation in animal models with OA.44 Kaempferol is a bioflavonoid with a variety of pharmacological activities, including anti-apoptotic and anti-inflammatory activities.37 Kaempferol has been reported to protect chondrocytes from lipopolysaccharide-induced inflammatory injury by down-regulating miR-146a.37 In addition, quercetin alleviates the progression of knee OA in rats by modulating TSC2-RHBE-mTOR signaling pathway.45 Our data further provide the evidence which supports CR is useful for ameliorating OA progression, via these bioactive components.
We also conducted bioinformatics analysis of 192 genes. The results showed that CR functioned through modulating multiple biological processes and pathways, mainly related to proteoglycans, PI3K-Akt signaling pathways, AGE-RAGE pathways and focal adhesion pathways. Proteoglycans are important components of the extracellular matrix of articular cartilage and provide vital biomechanical properties for its normal function. Evaluating proteoglycan molecules can be used as biomarkers of joint degradation in patients with OA, and some recent studies have reported that biomimetic proteoglycans strengthen the pericellular matrix of human cartilage, indicating targeting proteoglycans is a promising strategy to treat OA.46,47 The PI3K-Akt pathway and its downstream mTOR signaling are responsible for cartilage degeneration, synovial inflammation, subchondral bone sclerosis and osteophyte formation, so PI3K-Akt pathway inhibitors which block transduction of PI3K/AKT/mTOR signaling might be beneficial to OA treatment.48,49 Activation of RAGE signaling triggers the production of reactive oxygen species (ROS) via NADPH oxidase activation,45 and induces the activation of NF-κB, STAT3, AP-1, HIF-1α pathways, thus facilitating the inflammatory response.50 Activation of focal adhesion kinase (FAK) contributes to inflammatory injury of chondrocytes, and inhibition of FAK has been reported to attenuate OA in pre-clinical models.51 Comparing with an inhibitor targeting specific pathways, CRs have multiple bioactive components targeting multiple pathways, and may exert synergistic therapeutic effect for OA.
Furthermore, a series of core targets for CR in OA treatment were obtained, including AKT1, IL1B, SRC, BCL2, STAT3 and EGFR. AKT1, a member of the AKT kinase family, plays a key role in regulating apoptosis of chondrocytes.52 IL1B (expressing IL-1β, a pro-inflammatory cytokine) can inhibit chondrocyte viability and induce apoptosis.53 SRC is highly expressed in osteoclasts and can promote bone resorption, thus aggravating OA pathogenesis.54 Compared with healthy cartilage, the expression level of BCL2 in OA cartilage is reduced.55 STAT3 is a key transcription factor, involved in cartilage development.56 EGFR is highly expressed in healthy articular cartilage, and activation of EGFR signal can reduce the development of OA after load injury in mice.57 It is worth noting that we observed that, the key components of CR showed good binding affinity with AKT1, IL1B, SRC, BCL2, STAT3 and EGFR. Our findings partly explain the molecular mechanism by which CR exerts its anti-OA effect, via multiple bioactive ingredients. The degree values of the bioactive components in our analysis were similar (53, 53, 52, 52 for sugeonyl acetate, rosenonolactone, luteolin, kaempferol, respectively). Compared with luteolin and kaempferol, there are fewer reports about sugeonyl acetate and rosenonolactone, and their toxicology is unclear. Kaempferol was chosen for further investigation. Molecular dynamics simulation showed kaempferol interacted with AKT1, probably inhibiting its activation. This suggests that the regulatory effect of kaempferol on AKT signaling is a crucial mechanism by which CR protects chondrocytes. Chondrocytes are the main components of cartilage, and their dysfunction is related to the occurrence and development of OA.52 Pro-inflammatory cytokines secreted by chondrocytes, such as IL-1β and TNF-α, play a key role in the pathogenesis of OA.58 IL-1β has been reported to induce massive chondrocyte apoptosis and is widely used as an inducer of chondrocyte apoptosis in vitro.59 In this study, we found that chondrocyte viability was significantly reduced, and apoptosis and inflammation were aggravated when exposed to IL-1β, and kaempferol, a key component of CR, reversed the effect of IL-1β on chondrocyte. These experimental data further supported the protective role of CR on chondrocytes. However, the current work lacks the direct evidence to verify the binding relationship between the ingredients and the crucial targets, and necessary technologies such as HPLC-MS will further prove the regulatory of CR on these targets in the future. Additionally, the safety and anti-inflammatory effects of the other components, including sugeonyl acetate, rosenonolactone, luteolin and so on, need to be explored in the following work.
Of course, there are some other shortcomings in the present work. First of all, it’s worth noting that, the potential bioactive components of CR were retrieved from open access databases, and some potential functional components are probably missed. With the advancement of extraction, isolation and identification technologies of plant active ingredients, it is expected that more comprehensive pharmacological effects of CR will be elucidated in the future, which is of great significance to evaluate the therapeutic effect and potential side effects of CR. Secondly, even though network pharmacology is widely used to explain the pharmacological effect of herbal medicine, when screening the crucial components, topological analysis does not include the weight of a certain bioactive component of a herbal medicine in treating a disease. To be more specific, the relative contents of the bioactive components in CR are not considered in the analysis, while the components of herbs of different origin and seasons usually change. Therefore, the protective effects of other bioactive ingredients of CR on chondrocytes need to be verified in the following work, and quantification of these components in CR will help identify the crucial components and targets more precisely. Lastly, in vivo assays will further validate the bioactive components of CR in ameliorating chondrocytes injury and OA progression. Anyway, based on the available resources, this work tried to validate the potential value of CR in treating OA, and identified some promising natural monomers for drug development and clinical translation. Additionally, the binding affinity between the monomers and potential crucial targets was investigated (especially between kaempferol and AKT1). These findings lay the foundation of future studies.
Conclusion
Sugeonyl acetate, rosenonolactone, luteolin, kaempferol, chryseriol and quercetin may be the key components of CR in the treatment of OA. AKT1, IL1B, SRC, BCL2, STAT3, EGFR and so on were considered to be the key targets of CR in the treatment of OA. This study shows that CR plays a key role in the treatment of OA by acting on multiple targets and multiple pathways through multiple components. Our findings provide important information about the role of CR in OA treatment. However, it is worth noting that, its efficacy and safety remain to be validated by clinical trials before clinical application.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Ethics Statement
In the present work, no animal experiments are conducted, and no human tissues are used. This research was exempt from ethical approval, which was permitted by the Research Ethics Committee of The First Affiliated Hospital of Guangxi Traditional Chinese Medical University.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study is financially supported by the National Natural Science Fund of China (Grant No. 81860793) and the Guangxi Natural Science Foundation (Grant No.2020JJA140375).
Disclosure
The authors declare that they have no competing interests.
References
1. Ma L, Zhao X, Liu Y, Wu J, Yang X, Jin Q. Dihydroartemisinin attenuates osteoarthritis by inhibiting abnormal bone remodeling and angiogenesis in subchondral bone. Int J Mol Med. 2021;47(3):04855. doi:10.3892/ijmm.2021.4855
2. Sun T, Wang F, Hu G, Li Z. Salvianolic acid B activates chondrocytes autophagy and reduces chondrocyte apoptosis in obese mice via the KCNQ1OT1/miR-128-3p/SIRT1 signaling pathways. Nutr Metab. 2022;19(1):53. doi:10.1186/s12986-022-00686-0
3. Wang XY, Bao CC, An R, et al. Evaluation of the effect of physical therapy on pain and dysfunction of knee osteoarthritis based on fNIRS: a randomized controlled trial protocol. BMC Musculoskelet Disord. 2023;24(1):152. doi:10.1186/s12891-022-06074-2
4. Chen Z, Ge Y, Zhou L, et al. Pain relief and cartilage repair by Nanofat against osteoarthritis: preclinical and clinical evidence. Stem Cell Res Ther. 2021;12(1):477. doi:10.1186/s13287-021-02538-9
5. Cross M, Smith E, Hoy D, et al. The global burden of hip and knee osteoarthritis: estimates from the global burden of disease 2010 study. Ann Rheum Dis. 2014;73(7):1323–1330. doi:10.1136/annrheumdis-2013-204763
6. Bennell KL, Paterson KL, Metcalf BR, et al. Effect of intra-articular platelet-rich plasma vs placebo injection on pain and medial tibial cartilage volume in patients with knee osteoarthritis: the RESTORE randomized clinical trial. JAMA. 2021;326(20):2021–2030. doi:10.1001/jama.2021.19415
7. Sharma L. Osteoarthritis of the knee. N Engl J Med. 2021;384(1):51–59. doi:10.1056/NEJMcp1903768
8. Gunter BR, Butler KA, Wallace RL, Smith SM, Harirforoosh S. Non-steroidal anti-inflammatory drug-induced cardiovascular adverse events: a meta-analysis. J Clin Pharm Ther. 2017;42(1):27–38. doi:10.1111/jcpt.12484
9. An X, Wang R, Lv Z, et al. WTAP-mediated m6A modification of FRZB triggers the inflammatory response via the Wnt signaling pathway in osteoarthritis. Exp Mol Med. 2024;56(1):156–167.
10. Goldring MB, Otero M, Tsuchimochi K, Ijiri K, Li Y. Defining the roles of inflammatory and anabolic cytokines in cartilage metabolism. Ann Rheum Dis. 2008;67 Suppl 3(03):iii75–82.
11. Mogwasi R. Bio-accessibility of iron and copper from seven Kenyan anti-anaemia medicinal plants. Diag Therapeutics. 2022;2(1):1–12. doi:10.55976/dt.220231911-12
12. Zhu N, Hou J, Ma G, Liu J. Network pharmacology identifies the mechanisms of action of Shaoyao Gancao decoction in the treatment of osteoarthritis. Med Sci Monit. 2019;25:6051–6073. doi:10.12659/MSM.915821
13. Xiang C, Liao Y, Chen Z, et al. Network pharmacology and molecular docking to elucidate the potential mechanism of ligusticum chuanxiong against osteoarthritis. Front Pharmacol. 2022;13:854215. doi:10.3389/fphar.2022.854215
14. Wang F, Zhang S, Zhang J, Yuan F. Systematic review of ethnomedicine, phytochemistry, and pharmacology of Cyperi Rhizoma. Front Pharmacol. 2022;13:965902. doi:10.3389/fphar.2022.965902
15. Kanagali SN, Patil BM, Khanal P, Unger BS. Cyperus rotundus L. reverses the olanzapine-induced weight gain and metabolic changes-outcomes from network and experimental pharmacology. Comput Biol Med. 2022;141:105035. doi:10.1016/j.compbiomed.2021.105035
16. Kamala A, Middha SK, Karigar CS. Plants in traditional medicine with special reference to Cyperus rotundus L.: a review. 3 Biotech. 2018;8(7):309. doi:10.1007/s13205-018-1328-6
17. Xue BX, He RS, Lai JX, Mireku-Gyimah NA, Zhang LH, Wu HH. Phytochemistry, data mining, pharmacology, toxicology and the analytical methods of Cyperus rotundus L. (Cyperaceae): a comprehensive review. Phytochem Rev. 2023;15:1–46.
18. Mohamed AI, Beseni BK, Msomi NZ, et al. The antioxidant and antidiabetic potentials of polyphenolic-rich extracts of Cyperus rotundus (Linn.). J Biomol Struct Dyn. 2022;40(22):12075–12087. doi:10.1080/07391102.2021.1967197
19. Shakerin Z, Esfandiari E, Razavi S, Alaei H, Ghanadian M, Dashti G. Effects of Cyperus rotundus extract on spatial memory impairment and neuronal differentiation in rat model of Alzheimer’s disease. Adv Biomed Res. 2020;9:17. doi:10.4103/abr.abr_173_19
20. Zhang H, Li S, Lu J, et al. α-Cyperone (CYP) down-regulates NF-κB and MAPKs signaling, attenuating inflammation and extracellular matrix degradation in chondrocytes, to ameliorate osteoarthritis in mice. Aging. 2021;13(13):17690–17706. doi:10.18632/aging.203259
21. Berger SI, Iyengar R. Network analyses in systems pharmacology. Bioinformatics. 2009;25(19):2466–2472. doi:10.1093/bioinformatics/btp465
22. Xu X, Zhang Z, Liu L, Che C, Li W. Exploring the antiovarian cancer mechanisms of Salvia miltiorrhiza Bunge by network pharmacological analysis and molecular docking. Comput Math Methods Med. 2022;2022:7895246. doi:10.1155/2022/7895246
23. Wang Y, Xiao J, Suzek TO, et al. PubChem’s BioAssay database. Nucleic Acids Res. 2012;40(Database issue):D400–12. doi:10.1093/nar/gkr1132
24. Lin R, Lin X, Wu J, Chen T, Huang Z. Inhibitory Effects of Rabdosia rubescens in esophageal squamous cell carcinoma: network pharmacology and experimental validation. Evid Based Complement Alternat Med. 2022;2022:2696347. doi:10.1155/2022/2696347
25. Gfeller D, Michielin O, Zoete V. Shaping the interaction landscape of bioactive molecules. Bioinformatics. 2013;29(23):3073–3079. doi:10.1093/bioinformatics/btt540
26. Amberger JS, Hamosh A. Searching online Mendelian inheritance in man (OMIM): a knowledgebase of human genes and genetic phenotypes. Curr Protoc Bioinfo. 2017;58:1.2.1–1.2.12. doi:10.1002/cpbi.27
27. Safran M, Dalah I, Alexander J, et al. GeneCards Version 3: the human gene integrator. Database. 2010;2010:baq020. doi:10.1093/database/baq020
28. Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48(D1):D845–D855.
29. Dennis GJ, Sherman BT, Hosack DA, et al. Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):P3. doi:10.1186/gb-2003-4-5-p3
30. Szklarczyk D, Kirsch R, Koutrouli M, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51(D1):D638–D646. doi:10.1093/nar/gkac1000
31. Seeliger D, de Groot BL. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des. 2010;24(5):417–422. doi:10.1007/s10822-010-9352-6
32. Burley SK, Bhikadiya C, Bi C, et al. RCSB protein data bank (RCSB.org): delivery of experimentally-determined PDB structures alongside one million computed structure models of proteins from artificial intelligence/machine learning. Nucleic Acids Res. 2023;51(D1):D488–D508. doi:10.1093/nar/gkac1077
33. O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: an open chemical toolbox. J Cheminform. 2011;3:33. doi:10.1186/1758-2946-3-33
34. Morris GM, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi:10.1002/jcc.21256
35. Li Y, Duan J, Lin W, Liu J. Exosomal miR-93-5p regulated the progression of osteoarthritis by targeting ADAMTS9. Open Med. 2023;18(1):20230668. doi:10.1515/med-2023-0668
36. Zhuang Z, Ye G, Huang B. Kaempferol alleviates the interleukin-1β-induced inflammation in rat osteoarthritis chondrocytes via suppression of NF-κB. Med Sci Monit. 2017;23:3925–3931. doi:10.12659/MSM.902491
37. Saikia S, Bordoloi M. Molecular Docking: challenges, advances and its use in drug discovery perspective. Curr Drug Targets. 2019;20(5):501–521.
38. Jiang R, Hao P, Yu G, et al. Kaempferol protects chondrogenic ATDC5 cells against inflammatory injury triggered by lipopolysaccharide through down-regulating miR-146a. Int Immunopharmacol. 2019;69:373–381. doi:10.1016/j.intimp.2019.02.014
39. Wang G, Bi L, Li X, et al. Maintenance of effect of duloxetine in Chinese patients with pain due to osteoarthritis: 13-week open-label extension data. BMC Musculoskelet Disord. 2019;20(1):174. doi:10.1186/s12891-019-2527-y
40. Zhang J, Fan F, Liu A, et al. Icariin: a potential molecule for treatment of knee osteoarthritis. Front Pharmacol. 2022;13:811808. doi:10.3389/fphar.2022.811808
41. Zhang L, Shi X, Huang Z, et al. Network pharmacology approach to uncover the mechanism governing the effect of radix Achyranthis Bidentatae on osteoarthritis. BMC Complement Med Ther. 2020;20(1):121. doi:10.1186/s12906-020-02909-4
42. Dabaghian FH, Hashemi M, Entezari M, et al. Effect of Cyperus rotundus on ischemia-induced brain damage and memory dysfunction in rats. Iran J Basic Med Sci. 2015;18(2):199–204.
43. Babiaka SB, Moumbock AFA, Günther S, Ntie-Kang F. Natural products in Cyperus rotundus L. (Cyperaceae): an update of the chemistry and pharmacological activities. RSC Adv. 2021;11:15060–15077. doi:10.1039/D1RA00478F
44. Wang F, Song X, Ma S, et al. The treatment role of Cyperus rotundus L. to triple-negative breast cancer cells. Biosci Rep. 2019;39(6):BSR20190502. doi:10.1042/BSR20190502
45. Fei J, Liang B, Jiang C, Ni H, Wang L. Luteolin inhibits IL-1β-induced inflammation in rat chondrocytes and attenuates osteoarthritis progression in a rat model. Biomed Pharmacother. 2019;109:1586–1592. doi:10.1016/j.biopha.2018.09.161
46. Alcaide-Ruggiero L, Cugat R, Domínguez JM. Proteoglycans in articular cartilage and their contribution to chondral injury and repair mechanisms. Int J Mol Sci. 2023;24(13):10824. doi:10.3390/ijms241310824
47. Kahle ER, Fallahi H, Bergstrom AR, et al. Biomimetic proteoglycans strengthen the pericellular matrix of normal and osteoarthritic human cartilage. ACS Biomater Sci Eng. 2024;10(9):5617–5623. doi:10.1021/acsbiomaterials.4c00813
48. Sun K, Luo J, Guo J, Yao X, Jing X, Guo F. The PI3K/AKT/mTOR signaling pathway in osteoarthritis: a narrative review. Osteoarthritis Cartilage. 2020;28(4):400–409. doi:10.1016/j.joca.2020.02.027
49. Xue JF, Shi ZM, Zou J, Li XL. Inhibition of PI3K/AKT/mTOR signaling pathway promotes autophagy of articular chondrocytes and attenuates inflammatory response in rats with osteoarthritis. Biomed Pharmacother. 2017;89:1252–1261. doi:10.1016/j.biopha.2017.01.130
50. Rojas A, Lindner C, Schneider I, Gonzalez I, Uribarri J. The RAGE axis: a relevant inflammatory hub in human diseases. Biomolecules. 2024;14(4):412. doi:10.3390/biom14040412
51. Zhang C, Zhu M, Wang H, et al. LOXL2 attenuates osteoarthritis through inactivating integrin/FAK signaling. Sci Rep. 2021;11(1):17020. doi:10.1038/s41598-021-96348-x
52. Lv S, Wang X, Jin S, Shen S, Wang R, Tong P. Quercetin mediates TSC2-RHEB-mTOR pathway to regulate chondrocytes autophagy in knee osteoarthritis. Gene. 2022;820:146209. doi:10.1016/j.gene.2022.146209
53. Xue H, Tu Y, Ma T, et al. Lactoferrin inhibits IL-1β-induced chondrocyte apoptosis through AKT1-induced CREB1 activation. Cell Physiol Biochem. 2015;36(6):2456–2465. doi:10.1159/000430206
54. He L, Pan Y, Yu J, Wang B, Dai G, Ying X. Decursin alleviates the aggravation of osteoarthritis via inhibiting PI3K-Akt and NF-kB signal pathway. Int Immunopharmacol. 2021;97:107657. doi:10.1016/j.intimp.2021.107657
55. Feng WQ, Liu KY, Zhang JN, et al. Study on mechanism of Rehmanniae radix praeparata for treatment of osteoarthritis based on network pharmacology and molecular docking. Zhongguo Zhong Yao Za Zhi. 2022;47(19):5336–5343.
56. Wang BW, Jiang Y, Yao ZL, Chen PS, Yu B, Wang SN. Aucubin protects chondrocytes against IL-1β-induced apoptosis in vitro and inhibits osteoarthritis in mice model. Drug Des Devel Ther. 2019;13:3529–3538. doi:10.2147/DDDT.S210220
57. Shkhyan R, Van Handel B, Bogdanov J, et al. Drug-induced modulation of gp130 signalling prevents articular cartilage degeneration and promotes repair. Ann Rheum Dis. 2018;77(5):760–769. doi:10.1136/annrheumdis-2017-212037
58. Gui T, Wei Y, Luo L, et al. Activating EGFR signaling attenuates osteoarthritis development following loading injury in mice. J Bone Miner Res. 2022;37(12):2498–2511. doi:10.1002/jbmr.4717
59. Zheng X, Xia C, Chen Z, et al. Requirement of the phosphatidylinositol 3-kinase/Akt signaling pathway for the effect of nicotine on interleukin-1beta-induced chondrocyte apoptosis in a rat model of osteoarthritis. Biochem Biophys Res Commun. 2012;423(3):606–612. doi:10.1016/j.bbrc.2012.06.045
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, 3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
Recommended articles
Anti-Inflammatory Effects and Molecular Mechanisms of Shenmai Injection in Treating Acute Pancreatitis: Network Pharmacology Analysis and Experimental Verification
He Y, Hu C, Liu S, Xu M, Liang G, Du D, Liu T, Cai F, Chen Z, Tan Q, Deng L, Xia Q
Drug Design, Development and Therapy 2022, 16:2479-2495
Published Date: 2 August 2022
A Novel Approach Based on Gut Microbiota Analysis and Network Pharmacology to Explain the Mechanisms of Action of Cichorium intybus L. Formula in the Improvement of Hyperuricemic Nephropathy in Rats

Amatjan M, Li N, He P, Zhang B, Mai X, Jiang Q, Xie H, Shao X
Drug Design, Development and Therapy 2023, 17:107-128
Published Date: 20 January 2023
Network Pharmacology and Experimental Validation to Explore That Celastrol Targeting PTEN is the Potential Mechanism of Tripterygium wilfordii (Lév.) Hutch Against IgA Nephropathy

Zhao J, Liu H, Xia M, Chen Q, Wan L, Leng B, Tang C, Chen G, Liu Y, Zhang L, Liu H
Drug Design, Development and Therapy 2023, 17:887-900
Published Date: 23 March 2023
Investigating the Mechanism of Action of Schisandra chinensis Combined with Coenzyme Q10 in the Treatment of Heart Failure Based on PI3K-AKT Pathway

Wen S, Yang K, Bai Y, Wu Y, Liu D, Wu X, Zhang X, Sun J
Drug Design, Development and Therapy 2023, 17:939-957
Published Date: 27 March 2023
Network Pharmacology Analysis of the Mechanisms Underlying the Therapeutic Effects of Yangjing Zhongyu Tang on Thin Endometrium
Zhang L, Li H, Zhang L, Zu Z, Xu D, Zhang J
Drug Design, Development and Therapy 2023, 17:1805-1818
Published Date: 17 June 2023